Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation
- PMID: 21646403
- PMCID: PMC3115799
- DOI: 10.1083/jcb.201012035
Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation
Abstract
High expression of the protein kinase Bub1 has been observed in a variety of human tumors and often correlates with poor clinical prognosis, but its molecular and cellular consequences and role in tumorigenesis are unknown. Here, we demonstrate that overexpression of Bub1 in mice leads to near-diploid aneuploidies and tumor formation. We found that chromosome misalignment and lagging are the primary mitotic errors responsible for the observed aneuploidization. High Bub1 levels resulted in aberrant Bub1 kinase activity and hyperactivation of Aurora B kinase. When Aurora B activity is suppressed, pharmacologically or via BubR1 overexpression, chromosome segregation errors caused by Bub1 overexpression are largely corrected. Importantly, Bub1 transgenic mice overexpressing Bub1 developed various kinds of spontaneous tumors and showed accelerated Myc-induced lymphomagenesis. Our results establish that Bub1 has oncogenic properties and suggest that Aurora B is a critical target through which overexpressed Bub1 drives aneuploidization and tumorigenesis.
Figures

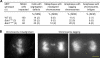
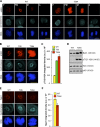
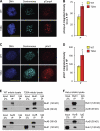
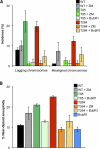
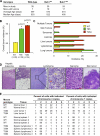
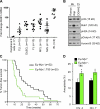
References
-
- Adams R.R., Maiato H., Earnshaw W.C., Carmena M. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880 10.1083/jcb.153.4.865 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

