miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1)
- PMID: 21646425
- PMCID: PMC3147603
- DOI: 10.1128/MCB.01433-10
miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1)
Abstract
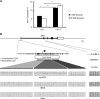
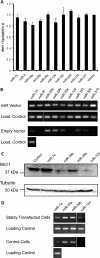
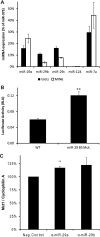
In pancreatic β cells, elevated glucose concentrations stimulate mitochondrial oxidative metabolism to raise intracellular ATP/ADP levels, prompting insulin secretion. Unusually low levels of expression of genes encoding the plasma membrane monocarboxylate transporter, MCT1 (SLC16A1), as well as lactate dehydrogenase A (LDHA) ensure that glucose-derived pyruvate is efficiently metabolized by mitochondria, while exogenous lactate or pyruvate is unable to stimulate metabolism and hence insulin secretion inappropriately. We show here that whereas DNA methylation at the Mct1 promoter is unlikely to be involved in cell-type-specific transcriptional repression, three microRNAs (miRNAs), miR-29a, miR-29b, and miR-124, selectively target both human and mouse MCT1 3' untranslated regions. Mutation of the cognate miR-29 or miR-124 binding sites abolishes the effects of the corresponding miRNAs, demonstrating a direct action of these miRNAs on the MCT1 message. However, despite reports of its expression in the mouse β-cell line MIN6, miR-124 was not detectably expressed in mature mouse islets. In contrast, the three isoforms of miR-29 are highly expressed and enriched in mouse islets. We show that inhibition of miR-29a in primary mouse islets increases Mct1 mRNA levels, demonstrating that miR-29 isoforms contribute to the β-cell-specific silencing of the MCT1 transporter and may thus affect insulin release.
Figures


References
-
- Baroukh N., et al. 2007. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J. Biol. Chem. 282:19575–19588 - PubMed
-
- Betel D., Wilson M., Gabow A., Marks D. S., Sander C. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res. 36:D149–D153 - PMC - PubMed
-
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6–21 - PubMed
-
- Bock C., et al. 2005. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21:4067–4068 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
