Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers
- PMID: 21646432
- PMCID: PMC3122625
- DOI: 10.1128/MMBR.00030-10
Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers
Abstract
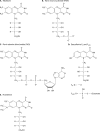
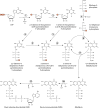
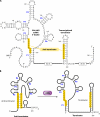
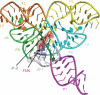
Riboflavin [7,8-dimethyl-10-(1'-d-ribityl)isoalloxazine, vitamin B₂] is an obligatory component of human and animal diets, as it serves as the precursor of flavin coenzymes, flavin mononucleotide, and flavin adenine dinucleotide, which are involved in oxidative metabolism and other processes. Commercially produced riboflavin is used in agriculture, medicine, and the food industry. Riboflavin synthesis starts from GTP and ribulose-5-phosphate and proceeds through pyrimidine and pteridine intermediates. Flavin nucleotides are synthesized in two consecutive reactions from riboflavin. Some microorganisms and all animal cells are capable of riboflavin uptake, whereas many microorganisms have distinct systems for riboflavin excretion to the medium. Regulation of riboflavin synthesis in bacteria occurs by repression at the transcriptional level by flavin mononucleotide, which binds to nascent noncoding mRNA and blocks further transcription (named the riboswitch). In flavinogenic molds, riboflavin overproduction starts at the stationary phase and is accompanied by derepression of enzymes involved in riboflavin synthesis, sporulation, and mycelial lysis. In flavinogenic yeasts, transcriptional repression of riboflavin synthesis is exerted by iron ions and not by flavins. The putative transcription factor encoded by SEF1 is somehow involved in this regulation. Most commercial riboflavin is currently produced or was produced earlier by microbial synthesis using special selected strains of Bacillus subtilis, Ashbya gossypii, and Candida famata. Whereas earlier RF overproducers were isolated by classical selection, current producers of riboflavin and flavin nucleotides have been developed using modern approaches of metabolic engineering that involve overexpression of structural and regulatory genes of the RF biosynthetic pathway as well as genes involved in the overproduction of the purine precursor of riboflavin, GTP.
Figures






References
-
- Abbas C., et al. March 2007. Transformation systems for flavinogenic yeast. U.S. patent 7,009,045
-
- Akiyama T., Selhub J., Rosenberg I. H. 1982. FMN phosphatase and FAD pyrophosphatase in rat intestinal brush borders: role in intestinal absorption of dietary riboflavin. J. Nutr. 112:263–268 - PubMed
-
- Alexopoulos C. J., Mims C. W., Blackwell M. 1996. Introductory mycology, 4th ed. John Wiley & Sons, Inc., New York, NY
-
- Ammelburg M., et al. 2007. A CTP-dependent archaeal riboflavin kinase forms a bridge in the evolution of cradle-loop barrels. Structure 15:1577–1590 - PubMed
-
- Asai S., Mase K., Yoshioka H. 2010. A key enzyme for flavin synthesis is required for nitric oxide and reactive oxygen species production in disease resistance. Plant J. 62:911–924 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

