Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift
- PMID: 21646514
- PMCID: PMC3121864
- DOI: 10.1073/pnas.1016719108
Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift
Abstract
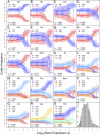
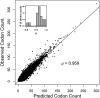
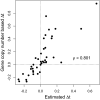
The genetic code is redundant with most amino acids using multiple codons. In many organisms, codon usage is biased toward particular codons. Understanding the adaptive and nonadaptive forces driving the evolution of codon usage bias (CUB) has been an area of intense focus and debate in the fields of molecular and evolutionary biology. However, their relative importance in shaping genomic patterns of CUB remains unsolved. Using a nested model of protein translation and population genetics, we show that observed gene level variation of CUB in Saccharomyces cerevisiae can be explained almost entirely by selection for efficient ribosomal usage, genetic drift, and biased mutation. The correlation between observed codon counts within individual genes and our model predictions is 0.96. Although a variety of factors shape patterns of CUB at the level of individual sites within genes, our results suggest that selection for efficient ribosome usage is a central force in shaping codon usage at the genomic scale. In addition, our model allows direct estimation of codon-specific mutation rates and elongation times and can be readily applied to any organism with high-throughput expression datasets. More generally, we have developed a natural framework for integrating models of molecular processes to population genetics models to quantitatively estimate parameters underlying fundamental biological processes, such a protein translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151:389–409. - PubMed
-
- Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. - PubMed
-
- Gilchrist MA. Combining models of protein translation and population genetics to predict protein production rates from codon usage patterns. Mol Biol Evol. 2007;24:2362–2372. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

