Preexisting drug-resistance mutations reveal unique barriers to resistance for distinct antivirals
- PMID: 21646519
- PMCID: PMC3121856
- DOI: 10.1073/pnas.1101515108
Preexisting drug-resistance mutations reveal unique barriers to resistance for distinct antivirals
Abstract
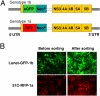
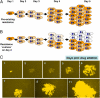
Clinical trials of direct-acting antiviral agents in patients chronically infected with hepatitis C virus (HCV) have demonstrated that viral resistance is detected rapidly during monotherapy. In patients, HCV does not exist as a single, genetically homogenous virus but rather as a population of variants termed "quasispecies." Preexisting variants resistant to specific antiviral drugs, overlooked in traditional hit-to-lead discovery efforts, may be responsible for these poor clinical outcomes. To enable real-time studies of resistance emergence in live cells, we established fluorescent protein-labeled HCV replicon cell lines. We validated these cell lines by demonstrating that antiviral susceptibility and the selection of signature resistance mutations for various drug classes are similar to traditional replicon cell lines. By quantifying the kinetics and uniformity of replication within colonies of drug-resistant fluorescent replicon cells, we showed that resistance emerged from a single cell and preexisted in a treatment-naive replicon population. Within this population, we determined the relative frequency of preexisting replicons capable of establishing foci during treatment with distinct antivirals. By measuring relative frequency as a function of dose, we quantitatively ranked distinct antiviral molecules on the basis of their distinct barriers to resistance. These insights into RNA virus quasispecies structure provide guidance for selecting clinical drug concentrations and selecting antiviral drug combinations most likely to suppress resistance.
Figures



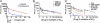
References
-
- Villano S, et al. Antiviral activity of the non-nucleoside polymerase inhibitor, HCV-796. Combination with PEGylated interferon alfa-2b in treatment-naive patients with chronic hepatitis C virus. J Hepatology. 2007;46(suppl):S24.
-
- Sarrazin C, Zeuzem S. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology. 2010;138:447–462. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

