Mimivirus shows dramatic genome reduction after intraamoebal culture
- PMID: 21646533
- PMCID: PMC3121840
- DOI: 10.1073/pnas.1101118108
Mimivirus shows dramatic genome reduction after intraamoebal culture
Erratum in
- Proc Natl Acad Sci U S A. 2011 Oct 11;108(41):17234
Abstract
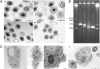
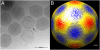
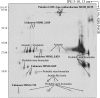
Most phagocytic protist viruses have large particles and genomes as well as many laterally acquired genes that may be associated with a sympatric intracellular life (a community-associated lifestyle with viruses, bacteria, and eukaryotes) and the presence of virophages. By subculturing Mimivirus 150 times in a germ-free amoebal host, we observed the emergence of a bald form of the virus that lacked surface fibers and replicated in a morphologically different type of viral factory. When studying a 0.40-μm filtered cloned particle, we found that its genome size shifted from 1.2 (M1) to 0.993 Mb (M4), mainly due to large deletions occurring at both ends of the genome. Some of the lost genes are encoding enzymes required for posttranslational modification of the structural viral proteins, such as glycosyltransferases and ankyrin repeat proteins. Proteomic analysis allowed identification of three proteins, probably required for the assembly of virus fibers. The genes for two of these were found to be deleted from the M4 virus genome. The proteins associated with fibers are highly antigenic and can be recognized by mouse and human antimimivirus antibodies. In addition, the bald strain (M4) was not able to propagate the sputnik virophage. Overall, the Mimivirus transition from a sympatric to an allopatric lifestyle was associated with a stepwise genome reduction and the production of a predominantly bald virophage resistant strain. The new axenic ecosystem allowed the allopatric Mimivirus to lose unnecessary genes that might be involved in the control of competitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

