IDH1 and IDH2 mutations in pediatric acute leukemia
- PMID: 21647154
- PMCID: PMC3883450
- DOI: 10.1038/leu.2011.133
IDH1 and IDH2 mutations in pediatric acute leukemia
Abstract
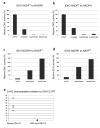
To investigate the frequency of isocitrate dehydrogenase 1 (IDH1) and 2 (IDH2) mutations in pediatric acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL), we sequenced these genes in diagnostic samples from 515 patients (227 AMLs and 288 ALLs). Somatic IDH1/IDH2 mutations were rare in ALL (N=1), but were more common in AML, occurring in 3.5% (IDH1 N=3 and IDH2 N=5), with the frequency higher in AMLs with a normal karyotype (9.8%). The identified IDH1 mutations occurred in codon 132 resulting in replacement of arginine with either cysteine (N=3) or histidine (N=1). By contrast, mutations in IDH2 did not affect the homologous residue but instead altered codon 140, resulting in replacement of arginine with either glutamine (N=4) or tryptophan (N=1). Structural modeling of IDH2 suggested that codon 140 mutations disrupt the enzyme's ability to bind its substrate isocitrate. Accordingly, recombinant IDH2 R140Q/W were unable to carry out the decarboxylation of isocitrate to α-ketoglutarate (α-KG), but instead gained the neomorphic activity to reduce α-KG to R(-)-2-hydroxyglutarete (2-HG). Analysis of primary leukemic blasts confirmed high levels of 2-HG in AMLs with IDH1/IDH2 mutations. Interestingly, 3/5 AMLs with IDH2 mutations had FLT3-activating mutations, raising the possibility that these mutations cooperate in leukemogenesis.
Figures
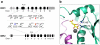
References
-
- Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia. Hematol Oncol Clin North Am. 2010;24:35–63. - PubMed
-
- Renneville A, Roumier C, Biggio V, Nibourel O, Boissel N, Fenaux P, et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22:915–931. - PubMed
-
- Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–2333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

