Discovery of protein phosphorylation motifs through exploratory data analysis
- PMID: 21647451
- PMCID: PMC3102080
- DOI: 10.1371/journal.pone.0020025
Discovery of protein phosphorylation motifs through exploratory data analysis
Abstract
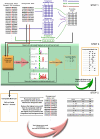
Background: The need for efficient algorithms to uncover biologically relevant phosphorylation motifs has become very important with rapid expansion of the proteomic sequence database along with a plethora of new information on phosphorylation sites. Here we present a novel unsupervised method, called Motif Finder (in short, F-Motif) for identification of phosphorylation motifs. F-Motif uses clustering of sequence information represented by numerical features that exploit the statistical information hidden in some foreground data. Furthermore, these identified motifs are then filtered to find "actual" motifs with statistically significant motif scores.
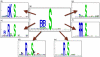
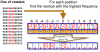
Results and discussion: We have applied F-Motif to several new and existing data sets and compared its performance with two well known state-of-the-art methods. In almost all cases F-Motif could identify all statistically significant motifs extracted by the state-of-the-art methods. More importantly, in addition to this, F-Motif uncovers several novel motifs. We have demonstrated using clues from the literature that most of these new motifs discovered by F-Motif are indeed novel. We have also found some interesting phenomena. For example, for CK2 kinase, the conserved sites appear only on the right side of S. However, for CDK kinase, the adjacent site on the right of S is conserved with residue P. In addition, three different encoding methods, including a novel position contrast matrix (PCM) and the simplest binary coding, are used and the ability of F-motif to discover motifs remains quite robust with respect to encoding schemes.
Conclusions: An iterative algorithm proposed here uses exploratory data analysis to discover motifs from phosphorylated data. The effectiveness of F-Motif has been demonstrated using several real data sets as well as using a synthetic data set. The method is quite general in nature and can be used to find other types of motifs also. We have also provided a server for F-Motif at http://f-motif.classcloud.org/, http://bio.classcloud.org/f-motif/ or http://ymu.classcloud.org/f-motif/.
Conflict of interest statement
Figures
References
-
- Pinna LA, Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta. 1996;1314:191–225. - PubMed
-
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. - PubMed
-
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. - PubMed
-
- Pawson T, Scott JD. Protein phosphorylation in signaling—50 years and counting. Trends Biochem Sci. 2005;30:286–290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

