Gene expression profile of the regeneration epithelium during axolotl limb regeneration
- PMID: 21648017
- PMCID: PMC3297817
- DOI: 10.1002/dvdy.22669
Gene expression profile of the regeneration epithelium during axolotl limb regeneration
Abstract
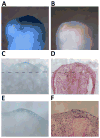
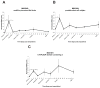
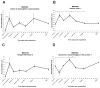
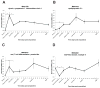
Urodele amphibians are unique among adult vertebrates in their ability to regenerate missing limbs. The process of limb regeneration requires several key tissues including a regeneration-competent wound epidermis called the regeneration epithelium (RE). We used microarray analysis to profile gene expression of the RE in the axolotl, a Mexican salamander. A list of 125 genes and expressed sequence tags (ESTs) showed a ≥1.5-fold expression in the RE than in a wound epidermis covering a lateral cuff wound. A subset of the RE ESTs and genes were further characterized for expression level changes over the time-course of regeneration. This study provides the first large scale identification of specific gene expression in the RE.
Copyright © 2011 Wiley-Liss, Inc.
Figures






References
-
- Bazzi H, Getz A, Mahoney MG, Ishida-Yamamoto A, Langbein L, Wahl JK, III, Christiano AM. Desmoglein 4 is expressed in highly differentiated keratinocytes and trichocytes in human epidermis and hair follicle. Differentiation. 2006;74:129–140. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300.
-
- Buchner G, Broccoli V, Bulfone A, Orfanelli U, Gattuso C, Ballabio A, Franco B. MAEG, an EGF-repeat containing gene, is a new marker associated with dermatome specification and morphogenesis of its derivatives. Mech Dev. 2000a;98:179–182. - PubMed
-
- Buchner G, Orfanelli U, Quaderi N, Bassi MT, Andolfi G, Ballabio A, Franco B. Identification of a New EGF-Repeat-Containing Gene from Human Xp22: A Candidate for Developmental Disorders. Genomics. 2000b;65:16–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

