Protein structure along the order-disorder continuum
- PMID: 21650183
- PMCID: PMC3125948
- DOI: 10.1021/ja203075p
Protein structure along the order-disorder continuum
Abstract
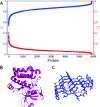
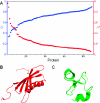
Thermal fluctuations cause proteins to adopt an ensemble of conformations wherein the relative stability of the different ensemble members is determined by the topography of the underlying energy landscape. "Folded" proteins have relatively homogeneous ensembles, while "unfolded" proteins have heterogeneous ensembles. Hence, the labels "folded" and "unfolded" represent attempts to provide a qualitative characterization of the extent of structural heterogeneity within the underlying ensemble. In this work, we introduce an information-theoretic order parameter to quantify this conformational heterogeneity. We demonstrate that this order parameter can be estimated in a straightforward manner from an ensemble and is applicable to both unfolded and folded proteins. In addition, a simple formula for approximating the order parameter directly from crystallographic B factors is presented. By applying these metrics to a large sample of proteins, we show that proteins span the full range of the order-disorder axis.
Figures

References
-
- Zweckstetter M.; Bax A. J. Am. Chem. Soc. 2000, 122, 3791.
-
- Salmon L.; Nodet G.; Ozenne V.; Yin G.; Jensen M. R.; Zweckstetter M.; Blackledge M. J. Am. Chem. Soc. 2010, 132, 8407. - PubMed
-
- Lange O. F.; Lakomek N. A.; Fares C.; Schroder G. F.; Walter K. F.; Becker S.; Meiler J.; Grubmuller H.; Griesinger C.; de Groot B. L. Science 2008, 320, 1471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

