The role of the Suppressor of Hairy-wing insulator protein in Drosophila oogenesis
- PMID: 21651900
- PMCID: PMC3143288
- DOI: 10.1016/j.ydbio.2011.05.666
The role of the Suppressor of Hairy-wing insulator protein in Drosophila oogenesis
Abstract
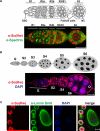
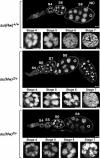
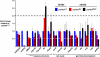
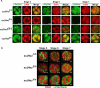
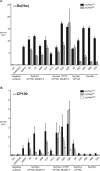
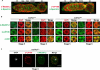
The Drosophila Suppressor of Hairy wing [Su(Hw)] insulator protein has an essential role in the development of the female germline. Here we investigate the function of Su(Hw) in the ovary. We show that Su(Hw) is universally expressed in somatic cells, while germ cell expression is dynamic. Robust levels accumulate in post-mitotic germ cells, where Su(Hw) localization is limited to chromosomes within nurse cells, the specialized cells that support oocyte growth. Although loss of Su(Hw) causes global defects in nurse cell chromosome structure, we demonstrate that these architectural changes are not responsible for the block in oogenesis. Connections between the fertility and insulator functions of Su(Hw) were investigated through studies of the two gypsy insulator proteins, Modifier of (mdg4)67.2 (Mod67.2) and Centrosomal Protein of 190kDa (CP190). Accumulation of these proteins is distinct from Su(Hw), with Mod67.2 and CP190 showing uniform expression in all cells during early stages of oogenesis that diminishes in later stages. Although Mod67.2 and CP190 extensively co-localize with Su(Hw) on nurse cell chromosomes, neither protein is required for nurse cell chromosome development or oocyte production. These data indicate that while the gypsy insulator function requires both Mod67.2 and CP190, these proteins are not essential for oogenesis. These studies represent the first molecular investigations of Su(Hw) function in the germline, which uncover distinct requirements for Su(Hw) insulator and ovary functions.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


References
-
- Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova GN, Lobanenkov VV, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol. 1997;17:1281–1288. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

