A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function
- PMID: 21652526
- PMCID: PMC3176121
- DOI: 10.1534/genetics.111.130781
A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function
Abstract
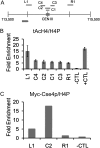
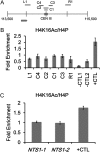
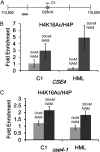
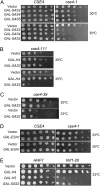
Hypoacetylated H4 is present at regional centromeres; however, its role in kinetochore function is poorly understood. We characterized H4 acetylation at point centromeres in Saccharomyces cerevisiae and determined the consequences of altered H4 acetylation on chromosome segregation. We observed low levels of tetra-acetylated and K16 acetylated histone H4 (H4K16Ac) at centromeres. Low levels of H4K16Ac were also observed at noncentromeric regions associated with Cse4p. Inhibition of histone deacetylases (HDAC) using nicotinamide (NAM) caused lethality in cse4 and hhf1-20 kinetochore mutants and increased centromeric H4K16Ac. Overexpression of Sas2-mediated H4K16 acetylation activity in wild-type cells led to increased rates of chromosome loss and synthetic dosage lethality in kinetochore mutants. Consistent with increased H4K16 acetylation as a cause of the phenotypes, deletion of the H4K16 deacetylase SIR2 or a sir2-H364Y catalytic mutant resulted in higher rates of chromosome loss compared to wild-type cells. Moreover, H4K16Q acetylmimic mutants displayed increased rates of chromosome loss compared to H4K16R nonacetylatable mutants and wild-type cells. Our work shows that hypoacetylated centromeric H4 is conserved across eukaryotic centromeres and hypoacetylation of H4K16 at centromeres plays an important role in accurate chromosome segregation.
Figures



References
-
- Bloom K. S., Carbon J., 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29: 305–317 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

