Arginine methylation of G3BP1 in response to Wnt3a regulates β-catenin mRNA
- PMID: 21652632
- PMCID: PMC3113675
- DOI: 10.1242/jcs.084046
Arginine methylation of G3BP1 in response to Wnt3a regulates β-catenin mRNA
Abstract
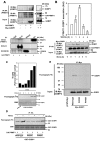
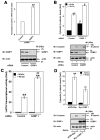
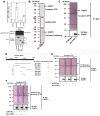
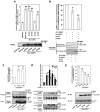
Wnt/β-catenin signaling is essential for normal mammalian development. Wnt3a activates the Wnt/β-catenin pathway through stabilization of β-catenin; a process in which the phosphoprotein Dishevelled figures prominently. Protein arginine methylation in signaling complexes containing Dishevelled was investigated. Mass spectrometry of a prominent arginine-methylated, Dishevelled-associated protein identified the Ras GTPase activating protein-binding protein 1 G3BP1. Stimulation of totipotent mouse embryonic F9 cells with Wnt3a provoked increased methylation of G3BP1. We show that G3BP1 is a novel Ctnnb1 mRNA binding protein. Methylation of G3BP1 constitutes a molecular switch that regulates Ctnnb1 mRNA in response to Wnt3a. Thus, the protein arginine methylation that targets G3BP1 acts as a novel regulator of Ctnnb1 mRNA.
Figures


References
-
- Aghib D. F., McCrea P. D. (1995). The E-cadherin complex contains the src substrate p120. Exp. Cell Res. 218, 359-369 - PubMed
-
- Angers S., Moon R. T. (2009). Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10, 468-477 - PubMed
-
- Atlas R., Behar L., Elliott E., Ginzburg I. (2004). The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 89, 613-626 - PubMed
-
- Atlas R., Behar L., Sapoznik S., Ginzburg I. (2007). Dynamic association with polysomes during P19 neuronal differentiation and an untranslated-region-dependent translation regulation of the tau mRNA by the tau mRNA-associated proteins IMP1, HuD, and G3BP1. J. Neurosci. Res. 85, 173-183 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

