The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton
- PMID: 21652697
- PMCID: PMC3143636
- DOI: 10.1074/jbc.M111.233700
The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton
Abstract
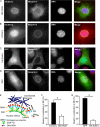
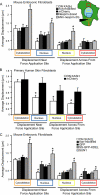
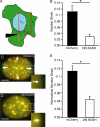
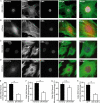
Maintaining physical connections between the nucleus and the cytoskeleton is important for many cellular processes that require coordinated movement and positioning of the nucleus. Nucleo-cytoskeletal coupling is also necessary to transmit extracellular mechanical stimuli across the cytoskeleton to the nucleus, where they may initiate mechanotransduction events. The LINC (Linker of Nucleoskeleton and Cytoskeleton) complex, formed by the interaction of nesprins and SUN proteins at the nuclear envelope, can bind to nuclear and cytoskeletal elements; however, its functional importance in transmitting intracellular forces has never been directly tested. This question is particularly relevant since recent findings have linked nesprin mutations to muscular dystrophy and dilated cardiomyopathy. Using biophysical assays to assess intracellular force transmission and associated cellular functions, we identified the LINC complex as a critical component for nucleo-cytoskeletal force transmission. Disruption of the LINC complex caused impaired propagation of intracellular forces and disturbed organization of the perinuclear actin and intermediate filament networks. Although mechanically induced activation of mechanosensitive genes was normal (suggesting that nuclear deformation is not required for mechanotransduction signaling) cells exhibited other severe functional defects after LINC complex disruption; nuclear positioning and cell polarization were impaired in migrating cells and in cells plated on micropatterned substrates, and cell migration speed and persistence time were significantly reduced. Taken together, our findings suggest that the LINC complex is critical for nucleo-cytoskeletal force transmission and that LINC complex disruption can result in defects in cellular structure and function that may contribute to the development of muscular dystrophies and cardiomyopathies.
Figures


References
-
- Padmakumar V. C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A. A., Gotzmann J., Foisner R., Karakesisoglou I. (2005) J. Cell Sci. 118, 3419–3430 - PubMed
-
- Padmakumar V. C., Abraham S., Braune S., Noegel A. A., Tunggal B., Karakesisoglou I., Korenbaum E. (2004) Exp. Cell Res. 295, 330–339 - PubMed
-
- Zhen Y. Y., Libotte T., Munck M., Noegel A. A., Korenbaum E. (2002) J. Cell Sci. 115, 3207–3222 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

