Protein kinase C (PKC) delta suppresses keratinocyte proliferation by increasing p21(Cip1) level by a KLF4 transcription factor-dependent mechanism
- PMID: 21652709
- PMCID: PMC3190685
- DOI: 10.1074/jbc.M110.205245
Protein kinase C (PKC) delta suppresses keratinocyte proliferation by increasing p21(Cip1) level by a KLF4 transcription factor-dependent mechanism
Retraction in
-
Withdrawal: Protein Kinase C (PKC) δ suppresses keratinocyte proliferation by increasing p21Cip1 level by a KLF4 transcription factor-dependent mechanism.J Biol Chem. 2025 Oct;301(10):110620. doi: 10.1016/j.jbc.2025.110620. Epub 2025 Sep 13. J Biol Chem. 2025. PMID: 40946414 Free PMC article. No abstract available.
Abstract
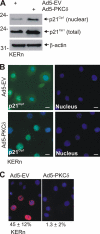
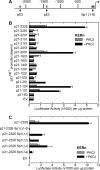
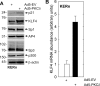
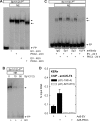
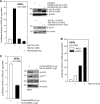
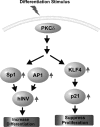
PKCδ increases keratinocyte differentiation and suppresses keratinocyte proliferation and survival. However, the mechanism of proliferation suppression is not well understood. The present studies show that PKCδ overexpression increases p21(Cip1) mRNA and protein level and promoter activity and that treatment with dominant-negative PKCδ, PKCδ-siRNA, or rottlerin inhibits promoter activation. Analysis of the p21(Cip1) promoter upstream regulatory region reveals three DNA segments that mediate PKCδ-dependent promoter activation. The PKCδ response element most proximal to the transcription start site encodes six GC-rich DNA elements. Mutation of these sites results in a loss of PKCδ-dependent promoter activation. Gel mobility supershift and chromatin immunoprecipitation reveal that these DNA elements bind the Kruppel-like transcription factor KLF4. PKCδ increases KLF4 mRNA and protein level and KLF4 binding to the GC-rich elements in the p21(Cip1) proximal promoter. In addition, KLF4-siRNA inhibits PKCδ-dependent p21(Cip1) promoter activity. PKCδ increases KLF4 expression leading to enhanced KLF4 interaction with the GC-rich elements in the p21(Cip1) promoter to activate transcription.
Figures




Comment in
-
Findings of Research Misconduct.Fed Regist. 2024 Aug 15;89(158):66420-66422. Fed Regist. 2024. PMID: 39161428 Free PMC article. No abstract available.
References
-
- Newton A. C. (1997) Curr. Opin. Cell Biol. 9, 161–167 - PubMed
-
- Nishizuka Y. (1992) Science 258, 607–614 - PubMed
-
- Rosse C., Linch M., Kermorgant S., Cameron A. J., Boeckeler K., Parker P. J. (2010) Nat. Rev. Mol. Cell Biol. 11, 103–112 - PubMed
-
- Gherzi R., Sparatore B., Patrone M., Sciutto A., Briata P. (1992) Biochem. Biophys. Res. Commun. 184, 283–291 - PubMed
-
- Matsui M. S., Chew S. L., DeLeo V. A. (1992) J. Invest. Dermatol. 99, 565–571 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

