G Protein binding sites on Calnuc (nucleobindin 1) and NUCB2 (nucleobindin 2) define a new class of G(alpha)i-regulatory motifs
- PMID: 21653697
- PMCID: PMC3151059
- DOI: 10.1074/jbc.M110.204099
G Protein binding sites on Calnuc (nucleobindin 1) and NUCB2 (nucleobindin 2) define a new class of G(alpha)i-regulatory motifs
Abstract
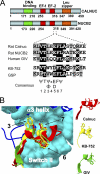
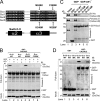
Heterotrimeric G proteins are molecular switches modulated by families of structurally and functionally related regulators. GIV (Gα-interacting vesicle-associated protein) is the first non-receptor guanine nucleotide exchange factor (GEF) that activates Gα(i) subunits via a defined, evolutionarily conserved motif. Here we found that Calnuc and NUCB2, two highly homologous calcium-binding proteins, share a common motif with GIV for Gα(i) binding and activation. Bioinformatics searches and structural analysis revealed that Calnuc and NUCB2 possess an evolutionarily conserved motif with sequence and structural similarity to the GEF sequence of GIV. Using in vitro pulldown and competition assays, we demonstrate that this motif binds preferentially to the inactive conformation of Gα(i1) and Gα(i3) over other Gα subunits and, like GIV, docks onto the α3/switch II cleft. Calnuc binding was impaired when Lys-248 in the α3 helix of Gα(i3) was replaced with M, the corresponding residue in Gα(o), which does not bind to Calnuc. Moreover, mutation of hydrophobic residues in the conserved motif predicted to dock on the α3/switch II cleft of Gα(i3) impaired the ability of Calnuc and NUCB2 to bind and activate Gα(i3) in vitro. We also provide evidence that calcium binding to Calnuc and NUCB2 abolishes their interaction with Gα(i3) in vitro and in cells, probably by inducing a conformational change that renders the Gα(i)-binding residues inaccessible. Taken together, our results identify a new type of Gα(i)-regulatory motif named the GBA motif (for Gα-binding and -activating motif), which is conserved across different proteins throughout evolution. These findings provide the structural basis for the properties of Calnuc and NUCB2 binding to Gα subunits and its regulation by calcium ions.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

