Cellular inhibitor of apoptosis protein-1 (cIAP1) can regulate E2F1 transcription factor-mediated control of cyclin transcription
- PMID: 21653699
- PMCID: PMC3143604
- DOI: 10.1074/jbc.M110.191239
Cellular inhibitor of apoptosis protein-1 (cIAP1) can regulate E2F1 transcription factor-mediated control of cyclin transcription
Abstract
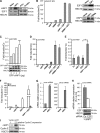
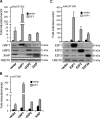
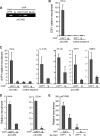
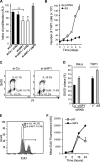
The inhibitor of apoptosis protein cIAP1 (cellular inhibitor of apoptosis protein-1) is a potent regulator of the tumor necrosis factor (TNF) receptor family and NF-κB signaling pathways in the cytoplasm. However, in some primary cells and tumor cell lines, cIAP1 is expressed in the nucleus, and its nuclear function remains poorly understood. Here, we show that the N-terminal part of cIAP1 directly interacts with the DNA binding domain of the E2F1 transcription factor. cIAP1 dramatically increases the transcriptional activity of E2F1 on synthetic and CCNE promoters. This function is not conserved for cIAP2 and XIAP, which are cytoplasmic proteins. Chromatin immunoprecipitation experiments demonstrate that cIAP1 is recruited on E2F binding sites of the CCNE and CCNA promoters in a cell cycle- and differentiation-dependent manner. cIAP1 silencing inhibits E2F1 DNA binding and E2F1-mediated transcriptional activation of the CCNE gene. In cells that express a nuclear cIAP1 such as HeLa, THP1 cells and primary human mammary epithelial cells, down-regulation of cIAP1 inhibits cyclin E and A expression and cell proliferation. We conclude that one of the functions of cIAP1 when localized in the nucleus is to regulate E2F1 transcriptional activity.
Figures




References
-
- Dubrez-Daloz L., Dupoux A., Cartier J. (2008) Cell Cycle 7, 1036–1046 - PubMed
-
- Gyrd-Hansen M., Meier P. (2010) Nat. Rev. Cancer 10, 561–574 - PubMed
-
- Varfolomeev E., Vucic D. (2008) Cell Cycle 7, 1511–1521 - PubMed
-
- Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) Cell 131, 669–681 - PubMed
-
- Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J. (2007) Cell 131, 682–693 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

