Functional dissection of the Oct6 Schwann cell enhancer reveals an essential role for dimeric Sox10 binding
- PMID: 21653862
- PMCID: PMC3137940
- DOI: 10.1523/JNEUROSCI.0659-11.2011
Functional dissection of the Oct6 Schwann cell enhancer reveals an essential role for dimeric Sox10 binding
Abstract
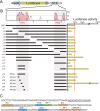
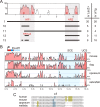
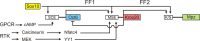
The POU domain transcription factor Pou3f1 (Oct6/Scip/Tst1) initiates the transition from ensheathing, promyelinating Schwann cells to myelinating cells. Axonal and other extracellular signals regulate Oct6 expression through the Oct6 Schwann cell enhancer (SCE), which is both required and sufficient to drive all aspects of Oct6 expression in Schwann cells. Thus, the Oct6 SCE is pivotal in the gene regulatory network that governs the onset of myelin formation in Schwann cells and provides a link between myelin promoting signaling and activation of a myelin-related transcriptional network. In this study, we define the relevant cis-acting elements within the SCE and identify the transcription factors that mediate Oct6 regulation. On the basis of phylogenetic comparisons and functional in vivo assays, we identify a number of highly conserved core elements within the mouse SCE. We show that core element 1 is absolutely required for full enhancer function and that it contains closely spaced inverted binding sites for Sox proteins. For the first time in vivo, the dimeric Sox10 binding to this element is shown to be essential for enhancer activity, whereas monomeric Sox10 binding is nonfunctional. As Oct6 and Sox10 synergize to activate the expression of the major myelin-related transcription factor Krox20, we propose that Sox10-dependent activation of Oct6 defines a feedforward regulatory module that serves to time and amplify the onset of myelination in the peripheral nervous system.
Figures



References
-
- Bansal R, Pfeiffer SE. Regulated galactolipid synthesis and cell surface expression in Schwann cell line D6P2T. J Neurochem. 1987;49:1902–1911. - PubMed
-
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. - PubMed
-
- Bermingham JR, Jr, Scherer SS, O'Connell S, Arroyo E, Kalla KA, Powell FL, Rosenfeld MG. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–1762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
