Trends of the major porin gene (ompF) evolution: insight from the genus Yersinia
- PMID: 21655186
- PMCID: PMC3105102
- DOI: 10.1371/journal.pone.0020546
Trends of the major porin gene (ompF) evolution: insight from the genus Yersinia
Abstract
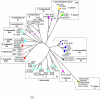
OmpF is one of the major general porins of Enterobacteriaceae that belongs to the first line of bacterial defense and interactions with the biotic as well as abiotic environments. Porins are surface exposed and their structures strongly reflect the history of multiple interactions with the environmental challenges. Unfortunately, little is known on diversity of porin genes of Enterobacteriaceae and the genus Yersinia especially. We analyzed the sequences of the ompF gene from 73 Yersinia strains covering 14 known species. The phylogenetic analysis placed most of the Yersinia strains in the same line assigned by 16S rDNA-gyrB tree. Very high congruence in the tree topologies was observed for Y. enterocolitica, Y. kristensenii, Y. ruckeri, indicating that intragenic recombination in these species had no effect on the ompF gene. A significant level of intra- and interspecies recombination was found for Y. aleksiciae, Y. intermedia and Y. mollaretii. Our analysis shows that the ompF gene of Yersinia has evolved with nonrandom mutational rate under purifying selection. However, several surface loops in the OmpF porin contain positively selected sites, which very likely reflect adaptive diversification Yersinia to their ecological niches. To our knowledge, this is a first investigation of diversity of the porin gene covering the whole genus of the family Enterobacteriaceae. This study demonstrates that recombination and positive selection both contribute to evolution of ompF, but the relative contribution of these evolutionary forces are different among Yersinia species.
Conflict of interest statement
Figures



References
-
- Sprague LD, Neubauer H. Yersinia aleksicieae sp. nov. Int J Syst Evol Microbiol. 2005;55:831–835. - PubMed
-
- Merhej V, Adekambi T, Pagnier I, Raoult D, Drancourt M. Yersinia massiliensis sp. nov., isolated from fresh water. Int J Syst Evol Microbiol. 2008;58:779–784. - PubMed
-
- Sprague LD, Scholz HC, Amann S, Busse HJ, Neubauer H. Yersinia similis sp. nov. Int J Syst Evol Microbiol. 2008;58:952–958. - PubMed
-
- Bottone EJ. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1999;1:323–333. - PubMed
-
- Sulakvelidze A. Yersinia other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis: the ignored species. Microbes Infect. 2000;2:497–513. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

