Dynamic Interaction of Hsp90 with Its Client Protein p53
- PMID: 21658391
- PMCID: PMC3143201
- DOI: 10.1016/j.jmb.2011.05.030
Dynamic Interaction of Hsp90 with Its Client Protein p53
Abstract
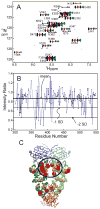
Although the structure of the molecular chaperone Hsp90 has been extensively characterized by X-ray crystallography, the nature of the interactions between Hsp90 and its client proteins remains unclear. We present results from a series of spectroscopic studies that strongly suggest that these interactions are highly dynamic in solution. Extensive NMR assignments have been made for human Hsp90 through the use of specific isotopic labeling of one- and two-domain constructs. Sites of interaction of a client protein, the p53 DNA-binding domain, were then probed both by chemical shift mapping and by saturation transfer NMR spectroscopy. Specific spectroscopic changes were small and difficult to observe, but were reproducibly measured for residues over a wide area of the Hsp90 surface in the N-terminal, middle and C-terminal domains. A somewhat greater specificity, for the area close to the interface between the N-terminal and middle domains of Hsp90, was identified in saturation transfer experiments. These results are consistent with a highly dynamic and nonspecific interaction between Hsp90 and p53 DNA-binding domain in this chaperone-client system, which results in changes in the client protein structure that are detectable by spectroscopic and other methods.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures







References
-
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. - PubMed
-
- Wegele H, Muller L, Buchner J. Hsp70 and Hsp90--a relay team for protein folding. Rev Physiol Biochem Pharmacol. 2004;151:1–44. - PubMed
-
- Hundley HA, Walter W, Bairstow S, Craig EA. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science. 2005;308:1032–1034. - PubMed
-
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. - PubMed
-
- Richter K, Buchner J. Hsp90: chaperoning signal transduction. J Cell Physiol. 2001;188:281–290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

