A SirT'N repression for Notch
- PMID: 21658598
- PMCID: PMC3491984
- DOI: 10.1016/j.molcel.2011.05.017
A SirT'N repression for Notch
Abstract
Mulligan et al. (2011) show here that the NAD(+)-dependent SIRT1 (H4K16; H1K26) deacetylase acts in concert with the LSD1 (H3K4) demethylase to repress Notch-induced transcription, thus coupling two distinct histone modifications at a key epigenetic switch for Notch target genes.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
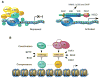
Comment on
-
A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development.Mol Cell. 2011 Jun 10;42(5):689-99. doi: 10.1016/j.molcel.2011.04.020. Epub 2011 May 19. Mol Cell. 2011. PMID: 21596603 Free PMC article.
References
-
- Bray S, Bernard F. Curr Top Dev Biol. 2010;92:253–275. - PubMed
-
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Cell. 2005;121:873–885. - PubMed
-
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP. Mol Cell. 2009;35:782–793. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

