Genome-scale analysis of aberrant DNA methylation in colorectal cancer
- PMID: 21659424
- PMCID: PMC3266034
- DOI: 10.1101/gr.117523.110
Genome-scale analysis of aberrant DNA methylation in colorectal cancer
Abstract
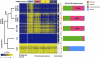
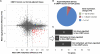
Colorectal cancer (CRC) is a heterogeneous disease in which unique subtypes are characterized by distinct genetic and epigenetic alterations. Here we performed comprehensive genome-scale DNA methylation profiling of 125 colorectal tumors and 29 adjacent normal tissues. We identified four DNA methylation-based subgroups of CRC using model-based cluster analyses. Each subtype shows characteristic genetic and clinical features, indicating that they represent biologically distinct subgroups. A CIMP-high (CIMP-H) subgroup, which exhibits an exceptionally high frequency of cancer-specific DNA hypermethylation, is strongly associated with MLH1 DNA hypermethylation and the BRAF(V600E) mutation. A CIMP-low (CIMP-L) subgroup is enriched for KRAS mutations and characterized by DNA hypermethylation of a subset of CIMP-H-associated markers rather than a unique group of CpG islands. Non-CIMP tumors are separated into two distinct clusters. One non-CIMP subgroup is distinguished by a significantly higher frequency of TP53 mutations and frequent occurrence in the distal colon, while the tumors that belong to the fourth group exhibit a low frequency of both cancer-specific DNA hypermethylation and gene mutations and are significantly enriched for rectal tumors. Furthermore, we identified 112 genes that were down-regulated more than twofold in CIMP-H tumors together with promoter DNA hypermethylation. These represent ∼7% of genes that acquired promoter DNA methylation in CIMP-H tumors. Intriguingly, 48/112 genes were also transcriptionally down-regulated in non-CIMP subgroups, but this was not attributable to promoter DNA hypermethylation. Together, we identified four distinct DNA methylation subgroups of CRC and provided novel insight regarding the role of CIMP-specific DNA hypermethylation in gene silencing.
Figures



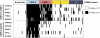
References
-
- Baylin SB, Ohm JE 2006. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6: 107–116 - PubMed
-
- Bernstein BE, Meissner A, Lander ES 2007. The mammalian epigenome. Cell 128: 669–681 - PubMed
-
- Bibikova M 2009. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics 1: 177–200 - PubMed
-
- Campan M, Weisenberger DJ, Trinh B, Laird PW 2009. MethyLight. Methods Mol Biol 507: 325–337 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
