Ligand-binding PAS domains in a genomic, cellular, and structural context
- PMID: 21663441
- PMCID: PMC3298442
- DOI: 10.1146/annurev-micro-121809-151631
Ligand-binding PAS domains in a genomic, cellular, and structural context
Abstract
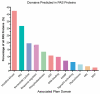
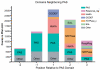
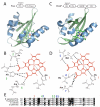
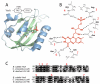
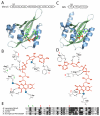
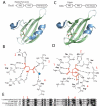
Per-Arnt-Sim (PAS) domains occur in proteins from all kingdoms of life. In the bacterial kingdom, PAS domains are commonly positioned at the amino terminus of signaling proteins such as sensor histidine kinases, cyclic-di-GMP synthases/hydrolases, and methyl-accepting chemotaxis proteins. Although these domains are highly divergent at the primary sequence level, the structures of dozens of PAS domains across a broad section of sequence space have been solved, revealing a conserved three-dimensional architecture. An all-versus-all alignment of 63 PAS structures demonstrates that the PAS domain family forms structural clades on the basis of two principal variables: (a) topological location inside or outside the plasma membrane and (b) the class of small molecule that they bind. The binding of a chemically diverse range of small-molecule metabolites is a hallmark of the PAS domain family. PAS ligand binding either functions as a primary cue to initiate a cellular signaling response or provides the domain with the capacity to respond to secondary physical or chemical signals such as gas molecules, redox potential, or photons. This review synthesizes the current state of knowledge of the structural foundations and evolution of ligand recognition and binding by PAS domains.
Figures





References
-
- Amezcua CA, Harper SM, Rutter J, Gardner KH. Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation. Structure. 2002;10:1349–61. - PubMed
-
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307:1271–92. - PubMed
-
- Anderson S, Crosson S, Moffat K. Short hydrogen bonds in photoactive yellow protein. Acta Crystallogr D Biol Crystallogr. 2004;60:1008–16. - PubMed
-
- Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

