Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye
- PMID: 21663797
- PMCID: PMC3117217
- DOI: 10.1016/j.cell.2011.05.003
Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye
Abstract
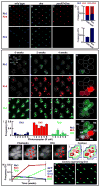
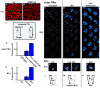
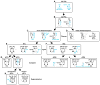
How complex networks of activators and repressors lead to exquisitely specific cell-type determination during development is poorly understood. In the Drosophila eye, expression patterns of Rhodopsins define at least eight functionally distinct though related subtypes of photoreceptors. Here, we describe a role for the transcription factor gene defective proventriculus (dve) as a critical node in the network regulating Rhodopsin expression. dve is a shared component of two opposing, interlocked feedforward loops (FFLs). Orthodenticle and Dve interact in an incoherent FFL to repress Rhodopsin expression throughout the eye. In R7 and R8 photoreceptors, a coherent FFL relieves repression by Dve while activating Rhodopsin expression. Therefore, this network uses repression to restrict and combinatorial activation to induce cell-type-specific expression. Furthermore, Dve levels are finely tuned to yield cell-type- and region-specific repression or activation outcomes. This interlocked FFL motif may be a general mechanism to control terminal cell-fate specification.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. - PubMed
-
- Briscoe AD. Reconstructing the ancestral butterfly eye: focus on the opsins. J Exp Biol. 2008;211:1805–1813. - PubMed
-
- Carr M, Hurley I, Fowler K, Pomiankowski A, Smith HK. Expression of defective proventriculus during head capsule development is conserved in Drosophila and stalk-eyed flies (Diopsidae) Dev Genes Evol. 2005;215:402–409. - PubMed
-
- Chou WH, Huber A, Bentrop J, Schulz S, Schwab K, Chadwell LV, Paulsen R, Britt SG. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development. 1999;126:607–616. - PubMed
-
- Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

