Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS)
- PMID: 21664291
- PMCID: PMC3199347
- DOI: 10.1016/j.bbalip.2011.05.007
Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS)
Abstract
Fundamental advances in secondary ion mass spectrometry (SIMS) now allow for the examination and characterization of lipids directly from biological materials. The successful application of SIMS-based imaging in the investigation of lipids directly from tissue and cells are demonstrated. Common complications and technical pitfalls are discussed. In this review, we examine the use of cluster ion sources and cryogenically compatible sample handling for improved ion yields and to expand the application potential of SIMS. Methodological improvements, including pre-treating the sample to improve ion yields and protocol development for 3-dimensional analyses (i.e. molecular depth profiling), are also included in this discussion. New high performance SIMS instruments showcasing the most advanced instrumental developments, including tandem MS capabilities and continuous ion beam compatibility, are described and the future direction for SIMS in lipid imaging is evaluated.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures

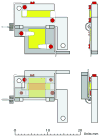



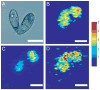

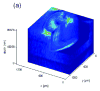

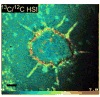




References
-
- Braun RM, Beyder A, Xu JY, Wood MC, Ewing AG, Winograd N. Spatially resolved detection of attomole quantities of organic molecules localized in picoliter vials using time-of-flight secondary ion mass spectrometry. Analytical Chemistry. 1999;71:3318–3324. - PubMed
-
- Kollmer F. Cluster primary ion bombardment of organic materials. Applied Surface Science. 2004;231:153–158.
-
- Fuchs B, Suss R, Schiller J. An update of MALDI-TOF mass spectrometry in lipid research. Prog Lipid Res. 2010;49:450–475. - PubMed
-
- Sullards MC, Chen Y, Liu Y, Merrill AH. Imaging of lipids directly from brain tissue via matrix assisted laser desorption ionization Time-of-Flight Mass Spectrometry (MALDI TOF MS) J Neurochem. 2010;113:92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

