Mitotic modulation of translation elongation factor 1 leads to hindered tRNA delivery to ribosomes
- PMID: 21665947
- PMCID: PMC3151038
- DOI: 10.1074/jbc.M111.255810
Mitotic modulation of translation elongation factor 1 leads to hindered tRNA delivery to ribosomes
Abstract
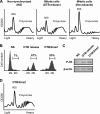
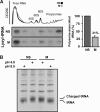
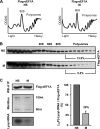
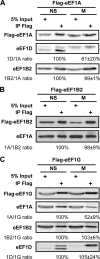
Translation elongation in eukaryotes is mediated by the concerted actions of elongation factor 1A (eEF1A), which delivers aminoacylated tRNA to the ribosome; elongation factor 1B (eEF1B) complex, which catalyzes the exchange of GDP to GTP on eEF1A; and eEF2, which facilitates ribosomal translocation. Here we present evidence in support of a novel mode of translation regulation by hindered tRNA delivery during mitosis. A conserved consensus phosphorylation site for the mitotic cyclin-dependent kinase 1 on the catalytic delta subunit of eEF1B (termed eEF1D) is required for its posttranslational modification during mitosis, resulting in lower affinity to its substrate eEF1A. This modification is correlated with reduced availability of eEF1A·tRNA complexes, as well as reduced delivery of tRNA to and association of eEF1A with elongating ribosomes. This mode of regulation by hindered tRNA delivery, although first discovered in mitosis, may represent a more globally applicable mechanism employed under other physiological conditions that involve down-regulation of protein synthesis at the elongation level.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

