Addition of genes for cellobiase and pectinolytic activity in Escherichia coli for fuel ethanol production from pectin-rich lignocellulosic biomass
- PMID: 21666025
- PMCID: PMC3147455
- DOI: 10.1128/AEM.05700-11
Addition of genes for cellobiase and pectinolytic activity in Escherichia coli for fuel ethanol production from pectin-rich lignocellulosic biomass
Abstract
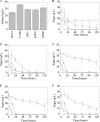
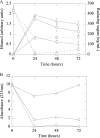
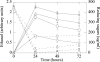
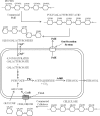
Ethanologenic Escherichia coli strain KO11 was sequentially engineered to contain the Klebsiella oxytoca cellobiose phosphotransferase genes (casAB) as well as a pectate lyase (pelE) from Erwinia chrysanthemi, yielding strains LY40A (casAB) and JP07 (casAB pelE), respectively. To obtain an effective secretion of PelE, the Sec-dependent pathway out genes from E. chrysanthemi were provided on a cosmid to strain JP07 to construct strain JP07C. Finally, oligogalacturonide lyase (ogl) from E. chrysanthemi was added to produce strain JP08C. E. coli strains LY40A, JP07, JP07C, and JP08C possessed significant cellobiase activity in cell lysates, while only strains JP07C and JP08C demonstrated extracellular pectate lyase activity. Fermentations conducted by using a mixture of pure sugars representative of the composition of sugar beet pulp (SBP) showed that strains LY40A, JP07, JP07C, and JP08C were able to ferment cellobiose, resulting in increased ethanol production from 15 to 45% in comparison to that of KO11. Fermentations with SBP at very low fungal enzyme loads during saccharification revealed significantly higher levels of ethanol production for LY40A, JP07C, and JP08C than for KO11. JP07C ethanol yields were not considerably higher than those of LY40A; however, oligogalacturonide polymerization studies showed an increased breakdown of biomass to small-chain (degree of polymerization, ≤6) oligogalacturonides. JP08C achieved a further breakdown of polygalacturonate to monomeric sugars, resulting in a 164% increase in ethanol yields compared to those of KO11. The addition of commercial pectin methylesterase (PME) further increased JP08C ethanol production compared to that of LY40A by demethylating the pectin for enzymatic attack by pectin-degrading enzymes.
Figures
References
-
- Atlas R. M. 1993. Handbook of microbiological media. CRC Press, Inc., Boca Raton, FL.
-
- Bao Y., Lies D. P., Fu H., Roberts G. P. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168 - PubMed
-
- Bradford M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 - PubMed
-
- Collmer A., Ried J. L., Mount M. S. 1988. Assay methods for pectic enzymes. Methods Enzymol. 161:329–335
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

