A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties
- PMID: 21666690
- PMCID: PMC5584938
- DOI: 10.1038/ni.2051
A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties
Abstract
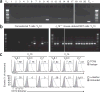
Type I natural killer T cells (NKT cells) are characterized by an invariant variable region 14-joining region 18 (V(α)14-J(α)18) T cell antigen receptor (TCR) α-chain and recognition of the glycolipid α-galactosylceramide (α-GalCer) restricted to the antigen-presenting molecule CD1d. Here we describe a population of α-GalCer-reactive NKT cells that expressed a canonical V(α)10-J(α)50 TCR α-chain, which showed a preference for α-glucosylceramide (α-GlcCer) and bacterial α-glucuronic acid-containing glycolipid antigens. Structurally, despite very limited TCRα sequence identity, the V(α)10 TCR-CD1d-α-GlcCer complex had a docking mode similar to that of type I TCR-CD1d-α-GalCer complexes, although differences at the antigen-binding interface accounted for the altered antigen specificity. Our findings provide new insight into the structural basis and evolution of glycolipid antigen recognition and have notable implications for the scope and immunological role of glycolipid-specific T cell responses.
Figures




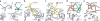
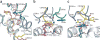
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

