Molecular basis of glyphosate resistance-different approaches through protein engineering
- PMID: 21668647
- PMCID: PMC3145815
- DOI: 10.1111/j.1742-4658.2011.08214.x
Molecular basis of glyphosate resistance-different approaches through protein engineering
Abstract
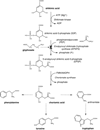
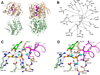
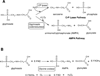
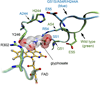
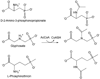
Glyphosate (N-phosphonomethyl-glycine) is the most widely used herbicide in the world: glyphosate-based formulations exhibit broad-spectrum herbicidal activity with minimal human and environmental toxicity. The extraordinary success of this simple, small molecule is mainly attributable to the high specificity of glyphosate for the plant enzyme enolpyruvyl shikimate-3-phosphate synthase in the shikimate pathway, leading to the biosynthesis of aromatic amino acids. Starting in 1996, transgenic glyphosate-resistant plants were introduced, thus allowing application of the herbicide to the crop (post-emergence) to remove emerged weeds without crop damage. This review focuses on mechanisms of resistance to glyphosate as obtained through natural diversity, the gene-shuffling approach to molecular evolution, and a rational, structure-based approach to protein engineering. In addition, we offer a rationale for the means by which the modifications made have had their intended effect.
© 2011 The Authors Journal compilation © 2011 FEBS.
Figures


References
-
- Padgette SR, Kolacz KH, Delannay X, Re DB, LaVallee BJ, Tinius CN, Rhodes WK, Otero YI, Barry GF, Eichholz DA, Peshke VM, Nida DL, Taylor NB, Kishore GM. Development, identification and characterization of a glyphosate-tolerant soybean line. Crop Science. 1995;35:1451–1461.
-
- Dill GM, CaJacob CA, Padgette SR. Glyphosate-resistant crops: adoption, use and future considerations. Pest Manag Sci. 2008;64:326–331. - PubMed
-
- Smith EA, Oehme FW. The biological activity of glyphosate to plants and animals: a literature review. Vet Hum Toxicol. 1992;34:531–543. - PubMed
-
- Williams GM, Kroes R, Munro IC. Safety evaluation and risk assessment of the herbicide Roundup and its active ingredient, glyphosate, for humans. Regul Toxicol Pharmacol. 2000;31:117–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

