Characterization of single-nucleotide variation in Indian-origin rhesus macaques (Macaca mulatta)
- PMID: 21668978
- PMCID: PMC3141668
- DOI: 10.1186/1471-2164-12-311
Characterization of single-nucleotide variation in Indian-origin rhesus macaques (Macaca mulatta)
Abstract
Background: Rhesus macaques are the most widely utilized nonhuman primate model in biomedical research. Previous efforts have validated fewer than 900 single nucleotide polymorphisms (SNPs) in this species, which limits opportunities for genetic studies related to health and disease. Extensive information about SNPs and other genetic variation in rhesus macaques would facilitate valuable genetic analyses, as well as provide markers for genome-wide linkage analysis and the genetic management of captive breeding colonies.
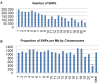
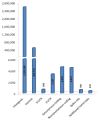
Results: We used the available rhesus macaque draft genome sequence, new sequence data from unrelated individuals and existing published sequence data to create a genome-wide SNP resource for Indian-origin rhesus monkeys. The original reference animal and two additional Indian-origin individuals were resequenced to low coverage using SOLiD™ sequencing. We then used three strategies to validate SNPs: comparison of potential SNPs found in the same individual using two different sequencing chemistries, and comparison of potential SNPs in different individuals identified with either the same or different sequencing chemistries. Our approach validated approximately 3 million SNPs distributed across the genome. Preliminary analysis of SNP annotations suggests that a substantial number of these macaque SNPs may have functional effects. More than 700 non-synonymous SNPs were scored by Polyphen-2 as either possibly or probably damaging to protein function and these variants now constitute potential models for studying functional genetic variation relevant to human physiology and disease.
Conclusions: Resequencing of a small number of animals identified greater than 3 million SNPs. This provides a significant new information resource for rhesus macaques, an important research animal. The data also suggests that overall genetic variation is high in this species. We identified many potentially damaging non-synonymous coding SNPs, providing new opportunities to identify rhesus models for human disease.
Figures


References
-
- Barr CS, Newman TK, Becker ML, Parker CC, Champoux M, Lesch KP, Goldman D, Suomi SJ, Higley JD. The utility of the non-human primate: model for studying gene by environment interactions in behavioral research. Genes Brain Behav. 2003;2(6):336–340. doi: 10.1046/j.1601-1848.2003.00051.x. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

