Chromosome-wide mapping of DNA methylation patterns in normal and malignant prostate cells reveals pervasive methylation of gene-associated and conserved intergenic sequences
- PMID: 21669002
- PMCID: PMC3124442
- DOI: 10.1186/1471-2164-12-313
Chromosome-wide mapping of DNA methylation patterns in normal and malignant prostate cells reveals pervasive methylation of gene-associated and conserved intergenic sequences
Abstract
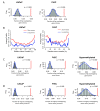
Background: DNA methylation has been linked to genome regulation and dysregulation in health and disease respectively, and methods for characterizing genomic DNA methylation patterns are rapidly emerging. We have developed/refined methods for enrichment of methylated genomic fragments using the methyl-binding domain of the human MBD2 protein (MBD2-MBD) followed by analysis with high-density tiling microarrays. This MBD-chip approach was used to characterize DNA methylation patterns across all non-repetitive sequences of human chromosomes 21 and 22 at high-resolution in normal and malignant prostate cells.
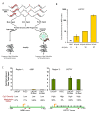
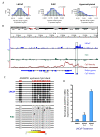
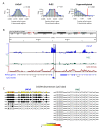
Results: Examining this data using computational methods that were designed specifically for DNA methylation tiling array data revealed widespread methylation of both gene promoter and non-promoter regions in cancer and normal cells. In addition to identifying several novel cancer hypermethylated 5' gene upstream regions that mediated epigenetic gene silencing, we also found several hypermethylated 3' gene downstream, intragenic and intergenic regions. The hypermethylated intragenic regions were highly enriched for overlap with intron-exon boundaries, suggesting a possible role in regulation of alternative transcriptional start sites, exon usage and/or splicing. The hypermethylated intergenic regions showed significant enrichment for conservation across vertebrate species. A sampling of these newly identified promoter (ADAMTS1 and SCARF2 genes) and non-promoter (downstream or within DSCR9, C21orf57 and HLCS genes) hypermethylated regions were effective in distinguishing malignant from normal prostate tissues and/or cell lines.
Conclusions: Comparison of chromosome-wide DNA methylation patterns in normal and malignant prostate cells revealed significant methylation of gene-proximal and conserved intergenic sequences. Such analyses can be easily extended for genome-wide methylation analysis in health and disease.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

