A Drosophila model for the Zellweger spectrum of peroxisome biogenesis disorders
- PMID: 21669930
- PMCID: PMC3180231
- DOI: 10.1242/dmm.007419
A Drosophila model for the Zellweger spectrum of peroxisome biogenesis disorders
Abstract
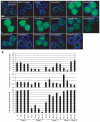
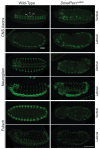
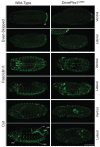
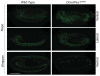
Human peroxisome biogenesis disorders are lethal genetic diseases in which abnormal peroxisome assembly compromises overall peroxisome and cellular function. Peroxisomes are ubiquitous membrane-bound organelles involved in several important biochemical processes, notably lipid metabolism and the use of reactive oxygen species for detoxification. Using cultured cells, we systematically characterized the peroxisome assembly phenotypes associated with dsRNA-mediated knockdown of 14 predicted Drosophila homologs of PEX genes (encoding peroxins; required for peroxisome assembly and linked to peroxisome biogenesis disorders), and confirmed that at least 13 of them are required for normal peroxisome assembly. We also demonstrate the relevance of Drosophila as a genetic model for the early developmental defects associated with the human peroxisome biogenesis disorders. Mutation of the PEX1 gene is the most common cause of peroxisome biogenesis disorders and is one of the causes of the most severe form of the disease, Zellweger syndrome. Inherited mutations in Drosophila Pex1 correlate with reproducible defects during early development. Notably, Pex1 mutant larvae exhibit abnormalities that are analogous to those exhibited by Zellweger syndrome patients, including developmental delay, poor feeding, severe structural abnormalities in the peripheral and central nervous systems, and early death. Finally, microarray analysis defined several clusters of genes whose expression varied significantly between wild-type and mutant larvae, implicating peroxisomal function in neuronal development, innate immunity, lipid and protein metabolism, gamete formation, and meiosis.
Figures






References
-
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., Amanatides P. G., Scherer S. E., Li P. W., Hoskins R. A., Galle R. F., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 - PubMed
-
- Bard F., Casano L., Mallabiabarrena A., Wallace E., Saito K., Kitayama H., Guizzunti G., Hu Y., Wendler F., DasGupta R., et al. (2006). Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439, 604–607 - PubMed
-
- Bilen J., Bonini N. M. (2005). Drosophila as a model for human neurodegenerative disease. Annu. Rev. Genet. 39, 153–171 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

