Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor
- PMID: 21670291
- PMCID: PMC3127895
- DOI: 10.1073/pnas.1104306108
Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor
Abstract
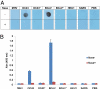
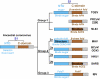
Coronaviruses have evolved diverse mechanisms to recognize different receptors for their cross-species transmission and host-range expansion. Mouse hepatitis coronavirus (MHV) uses the N-terminal domain (NTD) of its spike protein as its receptor-binding domain. Here we present the crystal structure of MHV NTD complexed with its receptor murine carcinoembryonic antigen-related cell adhesion molecule 1a (mCEACAM1a). Unexpectedly, MHV NTD contains a core structure that has the same β-sandwich fold as human galectins (S-lectins) and additional structural motifs that bind to the N-terminal Ig-like domain of mCEACAM1a. Despite its galectin fold, MHV NTD does not bind sugars, but instead binds mCEACAM1a through exclusive protein-protein interactions. Critical contacts at the interface have been confirmed by mutagenesis, providing a structural basis for viral and host specificities of coronavirus/CEACAM1 interactions. Sugar-binding assays reveal that galectin-like NTDs of some coronaviruses such as human coronavirus OC43 and bovine coronavirus bind sugars. Structural analysis and mutagenesis localize the sugar-binding site in coronavirus NTDs to be above the β-sandwich core. We propose that coronavirus NTDs originated from a host galectin and retained sugar-binding functions in some contemporary coronaviruses, but evolved new structural features in MHV for mCEACAM1a binding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



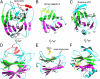
References
-
- Delmas B, Gelfi J, Sjöström H, Noren O, Laude H. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv Exp Med Biol. 1993;342:293–298. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

