Krüppel-like factor 4 regulates macrophage polarization
- PMID: 21670502
- PMCID: PMC3223832
- DOI: 10.1172/JCI45444
Krüppel-like factor 4 regulates macrophage polarization
Abstract
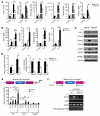
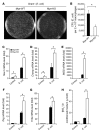
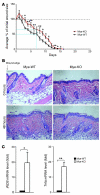
Current paradigms suggest that two macrophage subsets, termed M1 and M2, are involved in inflammation and host defense. While the distinct functions of M1 and M2 macrophages have been intensively studied - the former are considered proinflammatory and the latter antiinflammatory - the determinants of their speciation are incompletely understood. Here we report our studies that identify Krüppel-like factor 4 (KLF4) as a critical regulator of macrophage polarization. Macrophage KLF4 expression was robustly induced in M2 macrophages and strongly reduced in M1 macrophages, observations that were recapitulated in human inflammatory paradigms in vivo. Mechanistically, KLF4 was found to cooperate with Stat6 to induce an M2 genetic program and inhibit M1 targets via sequestration of coactivators required for NF-κB activation. KLF4-deficient macrophages demonstrated increased proinflammatory gene expression, enhanced bactericidal activity, and altered metabolism. Furthermore, mice bearing myeloid-specific deletion of KLF4 exhibited delayed wound healing and were predisposed to developing diet-induced obesity, glucose intolerance, and insulin resistance. Collectively, these data identify KLF4 as what we believe to be a novel regulator of macrophage polarization.
Figures
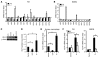

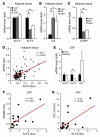

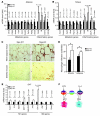
References
-
- Heilbronn LK, Campbell LV. Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Curr Pharm Des. 2008;14(12):1225–1230. - PubMed
-
- Charo IF. Macrophage polarization and insulin resistance: PPARgamma in control. Cell Metab. 2007;6(2):96–98. - PubMed
-
- Evans TJ. The role of macrophages in septic shock. Immunobiology. 1996;195(4–5):655–659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08HL083090/HL/NHLBI NIH HHS/United States
- K99/R00 HL097023/HL/NHLBI NIH HHS/United States
- R00 HL097023/HL/NHLBI NIH HHS/United States
- R01 HL086548/HL/NHLBI NIH HHS/United States
- R01 HL119195/HL/NHLBI NIH HHS/United States
- R01 HL110630/HL/NHLBI NIH HHS/United States
- HL086548/HL/NHLBI NIH HHS/United States
- R01 HL084154/HL/NHLBI NIH HHS/United States
- K08 HL083090/HL/NHLBI NIH HHS/United States
- K23 CA109115/CA/NCI NIH HHS/United States
- K99 HL097023/HL/NHLBI NIH HHS/United States
- R01 HL097593/HL/NHLBI NIH HHS/United States
- R01 HL075427/HL/NHLBI NIH HHS/United States
- HL075427/HL/NHLBI NIH HHS/United States
- HL097593/HL/NHLBI NIH HHS/United States
- HL076754/HL/NHLBI NIH HHS/United States
- 1 K23 CA109115-01A3/CA/NCI NIH HHS/United States
- HL084154/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

