Architecture of the flagellar rotor
- PMID: 21673656
- PMCID: PMC3160249
- DOI: 10.1038/emboj.2011.188
Architecture of the flagellar rotor
Abstract
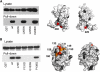
Rotation and switching of the bacterial flagellum depends on a large rotor-mounted protein assembly composed of the proteins FliG, FliM and FliN, with FliG most directly involved in rotation. The crystal structure of a complex between the central domains of FliG and FliM, in conjunction with several biochemical and molecular-genetic experiments, reveals the arrangement of the FliG and FliM proteins in the rotor. A stoichiometric mismatch between FliG (26 subunits) and FliM (34 subunits) is explained in terms of two distinct positions for FliM: one where it binds the FliG central domain and another where it binds the FliG C-terminal domain. This architecture provides a structural framework for addressing the mechanisms of motor rotation and direction switching and for unifying the large body of data on motor performance. Recently proposed alternative models of rotor assembly, based on a subunit contact observed in crystals, are not supported by experiment.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

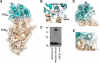



References
-
- Berg HC, Anderson RA (1973) Bacteria swim by rotating their flagellar filaments. Nature 245: 380–382 - PubMed
-
- Blair DF (2009) Structure and mechanism of the flagellar rotary motor. In Pili and Flagella: Current Research and Future Trends, Jarrell K (ed), 8, p 238 Caister Academic Press: Norfolk
-
- Blair DF, Berg HC (1988) Restoration of torque in defective flagellar motors. Science 242: 1678–1681 - PubMed
-
- Blair DF, Berg HC (1990) The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60: 439–449 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

