Colonization of Ireland: revisiting 'the pygmy shrew syndrome' using mitochondrial, Y chromosomal and microsatellite markers
- PMID: 21673740
- PMCID: PMC3242627
- DOI: 10.1038/hdy.2011.41
Colonization of Ireland: revisiting 'the pygmy shrew syndrome' using mitochondrial, Y chromosomal and microsatellite markers
Abstract
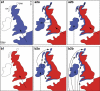
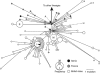
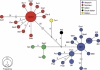
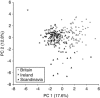
There is great uncertainty about how Ireland attained its current fauna and flora. Long-distance human-mediated colonization from southwestern Europe has been seen as a possible way that Ireland obtained many of its species; however, Britain has (surprisingly) been neglected as a source area for Ireland. The pygmy shrew has long been considered an illustrative model species, such that the uncertainty of the Irish colonization process has been dubbed 'the pygmy shrew syndrome'. Here, we used new genetic data consisting of 218 cytochrome (cyt) b sequences, 153 control region sequences, 17 Y-intron sequences and 335 microsatellite multilocus genotypes to distinguish between four possible hypotheses for the colonization of the British Isles, formulated in the context of previously published data. Cyt b sequences from western Europe were basal to those found in Ireland, but also to those found in the periphery of Britain and several offshore islands. Although the central cyt b haplotype in Ireland was found in northern Spain, we argue that it most likely occurred in Britain also, from where the pygmy shrew colonized Ireland as a human introduction during the Holocene. Y-intron and microsatellite data are consistent with this hypothesis, and the biological traits and distributional data of pygmy shrews argue against long-distance colonization from Spain. The compact starburst of the Irish cyt b expansion and the low genetic diversity across all markers strongly suggests a recent colonization. This detailed molecular study of the pygmy shrew provides a new perspective on an old colonization question.
Figures


References
-
- Balloux F, Ecoffey E, Fumagalli L, Goudet J, Wyttenbach A, Hausser J. Microsatellite conservation, polymorphism, and GC content in shrews of the genus Sorex (Insectivora, Mammalia) Mol Biol Evol. 1998;15:473–475. - PubMed
-
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. - PubMed
-
- Cassens I, Mardulyn P, Milinkovitch MC. Evaluating intraspecific ‘network' construction methods using simulated sequence data: do existing algorithms outperform the Global Maximum Parsimony approach. Syst Biol. 2005;54:363–372. - PubMed
-
- Churchfield S, Searle JB.2008aPygmy shrewIn: Harris S, Yalden DW (eds)Mammals of the British Isles Handbook4th edn.The Mammal Society: London; 267–271.
-
- Churchfield S, Searle JB.2008bCommon shrewIn: Harris S, Yalden DW (eds)Mammals of the British Isles Handbook4th edn,The Mammal Society: London; 257–265.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

