Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology
- PMID: 21673982
- PMCID: PMC3108599
- DOI: 10.1371/journal.pone.0020396
Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology
Abstract
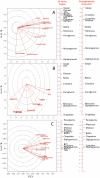
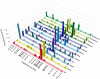
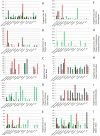
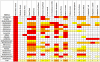
Endophytes are non-pathogenic microbes living inside plants. We asked whether endophytic species were conserved in the agriculturally important plant genus Zea as it became domesticated from its wild ancestors (teosinte) to modern maize (corn) and moved from Mexico to Canada. Kernels from populations of four different teosintes and 10 different maize varieties were screened for endophytic bacteria by culturing, cloning and DNA fingerprinting using terminal restriction fragment length polymorphism (TRFLP) of 16S rDNA. Principle component analysis of TRFLP data showed that seed endophyte community composition varied in relation to plant host phylogeny. However, there was a core microbiota of endophytes that was conserved in Zea seeds across boundaries of evolution, ethnography and ecology. The majority of seed endophytes in the wild ancestor persist today in domesticated maize, though ancient selection against the hard fruitcase surrounding seeds may have altered the abundance of endophytes. Four TRFLP signals including two predicted to represent Clostridium and Paenibacillus species were conserved across all Zea genotypes, while culturing showed that Enterobacter, Methylobacteria, Pantoea and Pseudomonas species were widespread, with γ-proteobacteria being the prevalent class. Twenty-six different genera were cultured, and these were evaluated for their ability to stimulate plant growth, grow on nitrogen-free media, solubilize phosphate, sequester iron, secrete RNAse, antagonize pathogens, catabolize the precursor of ethylene, produce auxin and acetoin/butanediol. Of these traits, phosphate solubilization and production of acetoin/butanediol were the most commonly observed. An isolate from the giant Mexican landrace Mixteco, with 100% identity to Burkholderia phytofirmans, significantly promoted shoot potato biomass. GFP tagging and maize stem injection confirmed that several seed endophytes could spread systemically through the plant. One seed isolate, Enterobacter asburiae, was able to exit the root and colonize the rhizosphere. Conservation and diversity in Zea-microbe relationships are discussed in the context of ecology, crop domestication, selection and migration.
Conflict of interest statement
Figures





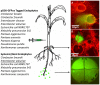
References
-
- Johnston-Monje D, Raizada MN. Plant and endophyte relationships: Nutrient management. In: Moo-Young M, editor. Comprehensive Biotechnology. 2 ed. Oxford: Elsevier; 2011.
-
- Kremer RJ. Identity and properties of bacteria inhabiting seeds of selected broadleaf weed species. Microb Ecol. 1987;14:29–37. - PubMed
-
- Schardl CL, Leuchtmann A, Spiering MJ. Symbioses of grasses with seedborne fungal endophytes. Annu Rev Plant Biol. 2004;55:315–340. - PubMed
-
- Dalling JW, Davis AS, Schutte BJ, Arnold AE. Seed survival in soil: interacting effects of predation, dormancy and the soil microbial community. J Ecol. 2011;99:89–95.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

