Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models
- PMID: 21674502
- PMCID: PMC3632393
- DOI: 10.1002/path.2891
Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models
Abstract
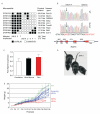
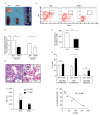
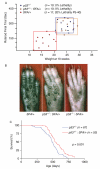
Ribosomal stress is an important, yet poorly understood, mechanism that results in activation of the p53 tumour suppressor. We present a mutation in the ribosomal protein Rpl27a gene (sooty foot ataxia mice), isolated through a sensitized N-ethyl-N-nitrosourea (ENU) mutagenesis screen for p53 pathway defects, that shares striking phenotypic similarities with high p53 mouse models, including cerebellar ataxia, pancytopenia and epidermal hyperpigmentation. This phenocopy is rescued in a haploinsufficient p53 background. A detailed examination of the bone marrow in these mice identified reduced numbers of haematopoietic stem cells and a p53-dependent c-Kit down-regulation. These studies suggest that reduced Rpl27a increases p53 activity in vivo, further evident with a delay in tumorigenesis in mutant mice. Taken together, these data demonstrate that Rpl27a plays a crucial role in multiple tissues and that disruption of this ribosomal protein affects both development and transformation.
Copyright © 2011 Pathological Society of Great Britain and Ireland. Published by John Wiley & Sons, Ltd.
Figures




References
-
- Chen D, Zhang Z, Li M, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

