Both innate and adaptive immunity mediate protective immunity against hepatitis C virus infection in chimpanzees
- PMID: 21674561
- PMCID: PMC3184181
- DOI: 10.1002/hep.24489
Both innate and adaptive immunity mediate protective immunity against hepatitis C virus infection in chimpanzees
Abstract
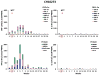
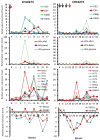
Understanding the immunological correlates associated with protective immunity following hepatitis C virus (HCV) reexposure is a prerequisite for the design of effective HCV vaccines and immunotherapeutics. In this study we performed a comprehensive analysis of innate and adaptive immunity following HCV reexposure of two chimpanzees that had previously recovered from HCV-JFH1 infection. One of the chimpanzees, CH10274, became protected from active viremia by repeated challenges with homologous HCV-JFH1 and developed neutralizing antibodies, but was later infected with high-level viremia by a heterologous challenge with the HCV H77 virus that persisted for more than 1 year. The other chimpanzee, CH10273, was protected from a similar, heterologous H77 challenge without any evidence of neutralizing antibodies. Peripheral HCV-specific T-cell responses were present in both chimpanzees after challenges and, interestingly, the overall magnitude of response was lower in uninfected CH10273, which, however, exhibited a more robust CD8+ T-cell response. CH10273 showed higher hepatic expression of CD8 and CD56 (natural killer) markers than CH10274 did shortly after inoculation with H77. The heightened T-cell response was associated with an enhanced hepatic production of interferons (both type I and II) and interferon-stimulated genes (ISGs) in CH10273. Therefore, protection or clearance of HCV reinfection upon heterologous rechallenge depends on the activation of both intrahepatic innate and cellular immune responses. Furthermore, our results suggest that serum neutralizing antibodies may contribute to early control of viral replication and spread after homologous HCV rechallenges but may not be sufficient for a long-term protective immunity.
Conclusion: Our study shows that protective immunity against HCV reinfection is orchestrated by a complex network of innate and adaptive immune responses.
Copyright © 2011 American Association for the Study of Liver Diseases.
Conflict of interest statement
Figures





References
-
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–29. - PubMed
-
- Argentini C, Genovese D, Dettori S, Rapicetta M. HCV genetic variability: from quasispecies evolution to genotype classification. Future Microbiol. 2009;4:359–373. - PubMed
-
- Gerlach JT, Diepolder HM, Zachoval R, Gruener NH, Jung MC, Ulsenheimer A, Schraut WW, et al. Acute hepatitis C: high rate of both spontaneous and treatment-induced viral clearance. Gastroenterology. 2003;125:80–88. - PubMed
-
- Grebely J, Conway B, Raffa JD, Lai C, Krajden M, Tyndall MW. Hepatitis C virus reinfection in injection drug users. Hepatology. 2006;44:1139–1145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
