Arp2/3 complex is bound and activated by two WASP proteins
- PMID: 21676863
- PMCID: PMC3158169
- DOI: 10.1073/pnas.1100236108
Arp2/3 complex is bound and activated by two WASP proteins
Abstract
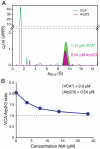
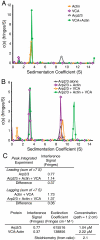
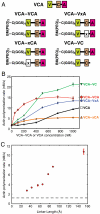
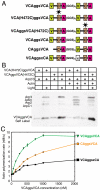
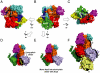
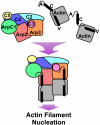
Actin related protein 2/actin related protein 3 (Arp2/3) complex nucleates new actin filaments in eukaryotic cells in response to signals from proteins in the Wiskott-Aldrich syndrome protein (WASP) family. The conserved VCA domain of WASP proteins activates Arp2/3 complex by inducing conformational changes and delivering the first actin monomer of the daughter filament. Previous models of activation have invoked a single VCA acting at a single site on Arp2/3 complex. Here we show that activation most likely involves engagement of two distinct sites on Arp2/3 complex by two VCA molecules, each delivering an actin monomer. One site is on Arp3 and the second is on ARPC1 and Arp2. The VCAs at these sites have distinct roles in activation. Our findings reconcile apparently conflicting literature on VCA activation of Arp2/3 complex and lead to a new model for this process.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. - PubMed
-
- Blanchoin L, et al. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. - PubMed
-
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

