Prediction of amyloid aggregation in vivo
- PMID: 21681200
- PMCID: PMC3128957
- DOI: 10.1038/embor.2011.116
Prediction of amyloid aggregation in vivo
Abstract
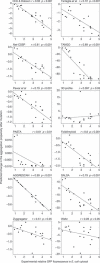
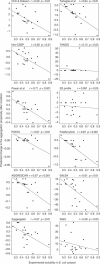
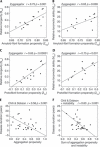
Many human diseases owe their pathology, to some degree, to the erroneous conversion of proteins from their soluble state into fibrillar, β-structured aggregates, often referred to as amyloid fibrils. Neurodegenerative diseases, such as Alzheimer and spongiform encephalopathies, as well as type 2 diabetes and both localized and systemic amyloidosis, are among the conditions that are associated with the formation of amyloid fibrils. Several mathematical tools can rationalize and even predict important parameters of amyloid fibril formation. It is not clear, however, whether such algorithms have predictive powers for in vivo systems, in which protein aggregation is affected by the presence of other biological factors. In this review, we briefly describe the existing algorithms and use them to predict the effects of mutations on the aggregation of specific proteins, for which in vivo experimental data are available. The comparison between the theoretical predictions and the experimental data obtained in vivo is shown for each algorithm and experimental data set, and statistically significant correlations are found in most cases.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Carrió M, González-Montalbán N, Vera A, Villaverde A, Ventura S (2005) Amyloid-like properties of bacterial inclusion bodies. J Mol Biol 347: 1025–1037 - PubMed
-
- Cherny I, Gazit E (2008) Amyloids: not only pathological agents but also ordered nanomaterials. Angew Chem Int Ed Engl 47: 4062–4069 - PubMed
-
- Chiti F, Dobson CM (2006) Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75: 333–366 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

