Transcriptional down-regulation of Brca1 and E-cadherin by CtBP1 in breast cancer
- PMID: 21681822
- PMCID: PMC3177013
- DOI: 10.1002/mc.20813
Transcriptional down-regulation of Brca1 and E-cadherin by CtBP1 in breast cancer
Abstract
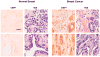
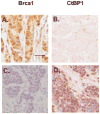
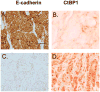
Carboxyl-terminal binding protein 1 (CtBP1) is a transcriptional co-repressor with oncogenic potential. Immunohistochemistry staining using human breast cancer tissue arrays revealed that 92% of invasive ductal breast cancer cases have CtBP1-positive staining compared to 4% CtBP1-positive in normal breast tissue. To explore the functional impact of CtBP1 in breast cancer, we examined CtBP1's transcriptional regulation of known tumor suppressors, breast cancer susceptibility gene 1 (Brca1), and E-cadherin. We found CtBP1 was recruited to the promoter regions of Brca1 and E-cadherin genes in breast cancer cells. Concomitantly, Brca1 loss was detected in 57% and E-cadherin loss was detected in 76% of human invasive ductal breast cancers, and correlated with CtBP1 nuclear staining in these lesions. Importantly, siRNA knock down of CtBP1 restored Brca1 and E-cadherin expression in breast cancer cell lines, implying CtBP1 down-regulates Brca1 and E-cadherin genes in human breast cancer. This study provides evidence that although genetic loss of Brca1 and E-cadherin are infrequent in breast cancer, they are down-regulated at the transcriptional level by CtBP1 expression. Thus, CtBP1 activation could be a potential biomarker for breast cancer development.
Copyright © 2011 Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare no conflict of interests.
Figures


References
-
- Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19(33):3823–3828. - PubMed
-
- Mroz EA, Baird AH, Michaud WA, Rocco JW. COOH-terminal binding protein regulates expression of the p16INK4A tumor suppressor and senescence in primary human cells. Cancer Res. 2008;68(15):6049–6053. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

