Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan
- PMID: 21682301
- PMCID: PMC3172152
- DOI: 10.1021/ja2040656
Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan
Abstract
The β-lactams are the most important class of antibiotics in clinical use. Their lethal targets are the transpeptidase domains of penicillin binding proteins (PBPs), which catalyze the cross-linking of bacterial peptidoglycan (PG) during cell wall synthesis. The transpeptidation reaction occurs in two steps, the first being formation of a covalent enzyme intermediate and the second involving attack of an amine on this intermediate. Here we use defined PG substrates to dissect the individual steps catalyzed by a purified E. coli transpeptidase. We demonstrate that this transpeptidase accepts a set of structurally diverse D-amino acid substrates and incorporates them into PG fragments. These results provide new information on donor and acceptor requirements as well as a mechanistic basis for previous observations that noncanonical D-amino acids can be introduced into the bacterial cell wall.
Figures

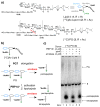


References
-
- Tipper DJ, Strominger JL. Proc Natl Acad Sci USA. 1965;54:1133–1141. - PMC - PubMed
- Ghuysen JM, Frere JM, Leyh-Bouille M, Coyette J, Dusart J, Nguyen-Disteche M. Annu Rev Biochem. 1979;48:73–101. - PubMed
- Waxman DJ, Strominger JL. Annu Rev Biochem. 1983;52:825–869. - PubMed
- Spratt BG. J Gen Microbiol. 1983;129:1247–1260. - PubMed
- Wilke MS, Lovering AL, Strynadka NC. Curr Opin Microbiol. 2005;8:525–533. - PubMed
- Testero SA, Fisher JF, Mobashery S. β-Lactam Antibiotics. In Burger’s Medicinal Chemistry. In: Abraham DJ, Rotella DP, editors. Drug Discovery and Development. Vol. 7. Wiley & Sons; New York: 2010. pp. 259–404.
-
-
Natural substrates refer to PG (or PG fragments) rather than beta-lactams (see reference 2) or modified peptides (see reference 10). See the following for studies that utilize PG substrates: Waxman DJ, Yu W, Strominger JL. J Biol Chem. 1980;255:11577–11587.Schwartz B, Markwalder JA, Wang Y. J Am Chem Soc. 2001;123:11638–11643.Bertsche U, Breukink E, Kast T, Vollmer W. J Biol Chem. 2005;280:38096–38101.Born P, Breukink E, Vollmer W. J Biol Chem. 2006;281:26985–26993.
-
-
-
The TP domains discussed in this work are fused to N-terminal PGT domains in bifunctional proteins. These TPs had not been shown to catalyze PG hydrolysis using natural substrates. For a review, see: Goffin C, Ghuysen JM. Microbiol Mol Biol Rev. 2002;66:702–738.
-
-
-
For many Gram-positive bacteria, the epsilon-amino group of L-Lys is acylated with other amino acids or peptides, which are involved in cross-links. See the following for a review on PG structure: Vollmer W, Blanot D, de Pedro MA. FEMS Microbiol Rev. 2008;32:149–167.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

