Blocking premature reverse transcription fails to rescue the HIV-1 nucleocapsid-mutant replication defect
- PMID: 21682883
- PMCID: PMC3141651
- DOI: 10.1186/1742-4690-8-46
Blocking premature reverse transcription fails to rescue the HIV-1 nucleocapsid-mutant replication defect
Abstract
Background: The nucleocapsid (NC) protein of HIV-1 is critical for viral replication. Mutational analyses have demonstrated its involvement in viral assembly, genome packaging, budding, maturation, reverse transcription, and integration. We previously reported that two conservative NC mutations, His23Cys and His44Cys, cause premature reverse transcription such that mutant virions contain approximately 1,000-fold more DNA than wild-type virus, and are replication defective. In addition, both mutants show a specific defect in integration after infection.
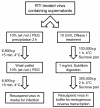
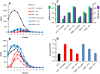
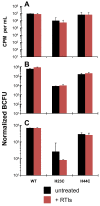
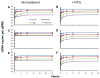
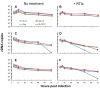
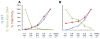
Results: In the present study we investigated whether blocking premature reverse transcription would relieve the infectivity defects, which we successfully performed by transfecting proviral plasmids into cells cultured in the presence of high levels of reverse transcriptase inhibitors. After subsequent removal of the inhibitors, the resulting viruses showed no significant difference in single-round infective titer compared to viruses where premature reverse transcription did occur; there was no rescue of the infectivity defects in the NC mutants upon reverse transcriptase inhibitor treatment. Surprisingly, time-course endogenous reverse transcription assays demonstrated that the kinetics for both the NC mutants were essentially identical to wild-type when premature reverse transcription was blocked. In contrast, after infection of CD4+ HeLa cells, it was observed that while the prevention of premature reverse transcription in the NC mutants resulted in lower quantities of initial reverse transcripts, the kinetics of reverse transcription were not restored to that of untreated wild-type HIV-1.
Conclusions: Premature reverse transcription is not the cause of the replication defect but is an independent side-effect of the NC mutations.
Figures

References
-
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

