Novel norovirus recombinants and of GII.4 sub-lineages associated with outbreaks between 2006 and 2010 in Belgium
- PMID: 21682917
- PMCID: PMC3135559
- DOI: 10.1186/1743-422X-8-310
Novel norovirus recombinants and of GII.4 sub-lineages associated with outbreaks between 2006 and 2010 in Belgium
Abstract
Background: Noroviruses (NoVs) are an important cause of acute gastroenteritis in humans worldwide. To gain insight into the epidemiologic patterns of NoV outbreaks and to determine the genetic variation of NoVs strains circulating in Belgium, stool samples originating from patients infected with NoVs in foodborne outbreak investigations were analysed between December 2006 and December 2010.
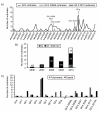
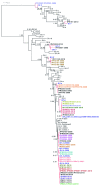
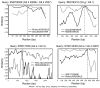
Results: NoVs were found responsible of 11.8% of all suspected foodborne outbreaks reported in the last 4 years and the number of NoV outbreaks reported increased along the years representing more than 30% of all foodborne outbreaks in 2010. Genogroup II outbreaks largely predominated and represented more than 90% of all outbreaks. Phylogenetic analyses were performed with 63 NoV-positive samples for the partial polymerase (N = 45) and/or capsid gene (N = 35) sequences. For 12 samples, sequences covering the ORF1-ORF2 junction were obtained. A variety of genotypes was found among genogroups I and II; GII.4 was predominant followed in order of importance by GII.2, GII.7, GII.13, GI.4 and GI.7. In the study period, GII.4 NoVs variants 2006a, 2006b, 2007, 2008 and 2010 were identified. Moreover, phylogenetic analyses identified different recombinant NoV strains that were further characterised as intergenotype (GII.e/GII.4 2007, GII.e/GII.3 and GII.g/GII.1) and intersub-genotype (GII.4 2006b/GII.4 2007 and GII.4 2010/GII.4 2010b) recombinants.
Conclusions: NoVs circulating in the last 4 years in Belgium showed remarkable genetic diversity either by small-scale mutations or genetic recombination. In this period, GII.4 2006b was successfully displaced by the GII.4 2010 subtype, and previously reported epidemic GII.b recombinants seemed to have been superseded by GII.e recombinants in 2009 and GII.g recombinants in 2010. This study showed that the emergence of novel GII.4 variants together with novel GII recombinants could lead to an explosion in NoV outbreaks, likewise to what was observed in 2008 and 2010. Among recombinants detected in this study, two hitherto unreported strains GII.e/GII.3 and GII.g/GII.1 were characterised. Surveillance will remain important to monitor contemporaneously circulating strains in order to adapt preventive and curative strategies.
Figures

References
-
- Siebenga J, Duizer E, Koopmans M. In: Caliciviruses: Molecular and Cellular Virology. Hansman GS, Jiang XJ, Green KY, editor. Norfolk: Caister Academic Press; 2010. Norovirus Epidemiology; pp. 1–24.
-
- Leuenberger S, Widdowson M, Feilchenfeldt J, Egger R, Streuli R. Norovirus outbreak in a district general hospital - new strain identified. Swiss Med Wkly. 2007;137:57–81. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

