Using Caenorhabditis elegans to study serpinopathies
- PMID: 21683258
- PMCID: PMC4374434
- DOI: 10.1016/B978-0-12-386471-0.00013-4
Using Caenorhabditis elegans to study serpinopathies
Abstract
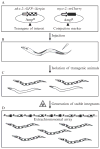
Protein misfolding, polymerization, and/or aggregation are hallmarks of serpinopathies and many other human genetic disorders including Alzheimer's, Huntington's, and Parkinson's disease. While higher organism models have helped shape our understanding of these diseases, simpler model systems, like Caenorhabditis elegans, offer great versatility for elucidating complex genetic mechanisms underlying these diseases. Moreover, recent advances in automated high-throughput methodologies have promoted C. elegans as a useful tool for drug discovery. In this chapter, we describe how one could model serpinopathies in C. elegans and how one could exploit this model to identify small molecule compounds that can be developed into effective therapeutic drugs.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



References
-
- Bleicher KH, et al. Hit and lead generation: Beyond high-throughput screening. Nat Rev Drug Discov. 2003;2(5):369–378. - PubMed
-
- Chalfie M, et al. Green fluorescent protein as a marker for gene expression. Science. 1994;263(5148):802–805. - PubMed
-
- Evans TC. WormBook, editor. The C. elegans Research Community. WormBook; 2006. Transformation and microinjection. http://www.wormbook.org. - DOI
-
- Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93(2):189–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

