Deciphering the RNA polymerase II CTD code in fission yeast
- PMID: 21684186
- PMCID: PMC3142328
- DOI: 10.1016/j.molcel.2011.05.024
Deciphering the RNA polymerase II CTD code in fission yeast
Abstract
The RNA polymerase II carboxy-terminal domain (CTD) consists of tandem Y(1)S(2)P(3)T(4)S(5)P(6)S(7) repeats. Dynamic remodeling of the CTD, especially its serine phosphorylation pattern, conveys informational cues about the transcription apparatus to a large ensemble of CTD-binding proteins. Our genetic dissection of fission yeast CTD function provides insights to the "CTD code." Two concepts stand out. First, the Ser2 requirement for transcription during sexual differentiation is bypassed by subtracting Ser7, signifying that imbalance in the phosphorylation array, not absence of a phospho-CTD cue, underlies a CTD-associated pathology. Second, the essentiality of Ser5 for vegetative growth is circumvented by covalently tethering mRNA capping enzymes to the CTD, thus proving that capping enzyme recruitment is a chief function of the Ser5-PO(4) mark. This illustrates that a key "letter" in the CTD code can be neutralized by delivering its essential cognate receptor to the transcription complex via an alternative route.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


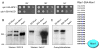
Comment in
-
Gimme phospho-serine five! Capping enzyme guanylyltransferase recognition of the RNA polymerase II CTD.Mol Cell. 2011 Jul 22;43(2):163-5. doi: 10.1016/j.molcel.2011.07.003. Mol Cell. 2011. PMID: 21777807
References
-
- Coudreuse D, van Bakel H, Dewez M, Soutourina J, Parnell T, Vandenhaute J, Cairns B, Werner M, Hermand D. A gene-specific requirement of RNA polymerase II CTD phosphorylation for sexual differentiation in S. pombe. Curr Biol. 2010;20:1053–1064. - PubMed
-
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genetics. 2008;24:280–288. - PubMed
-
- Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxyl-terminal domain of RNA polymerase II. Mol Cell. 2003;11:1549–1561. - PubMed
-
- Hausmann S, Koiwa H, Krishnamurthy S, Hampsey M, Shuman S. Different strategies for carboxyl-terminal domain (CTD) recognition by Serine5-specific CTD phosphatases. J Biol Chem. 2005;280:37681–37688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

