IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming
- PMID: 21685106
- PMCID: PMC3117384
- DOI: 10.1093/bioinformatics/btr215
IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming
Abstract
Motivation: Pseudoknots found in secondary structures of a number of functional RNAs play various roles in biological processes. Recent methods for predicting RNA secondary structures cover certain classes of pseudoknotted structures, but only a few of them achieve satisfying predictions in terms of both speed and accuracy.
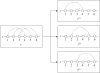
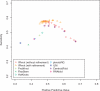
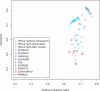
Results: We propose IPknot, a novel computational method for predicting RNA secondary structures with pseudoknots based on maximizing expected accuracy of a predicted structure. IPknot decomposes a pseudoknotted structure into a set of pseudoknot-free substructures and approximates a base-pairing probability distribution that considers pseudoknots, leading to the capability of modeling a wide class of pseudoknots and running quite fast. In addition, we propose a heuristic algorithm for refining base-paring probabilities to improve the prediction accuracy of IPknot. The problem of maximizing expected accuracy is solved by using integer programming with threshold cut. We also extend IPknot so that it can predict the consensus secondary structure with pseudoknots when a multiple sequence alignment is given. IPknot is validated through extensive experiments on various datasets, showing that IPknot achieves better prediction accuracy and faster running time as compared with several competitive prediction methods.
Availability: The program of IPknot is available at http://www.ncrna.org/software/ipknot/. IPknot is also available as a web server at http://rna.naist.jp/ipknot/.
Contact: satoken@k.u-tokyo.ac.jp; ykato@is.naist.jp
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
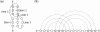



References
-
- Akutsu T. Dynamic programming algorithms for RNA secondary structure prediction with pseudoknots. Discrete Appl. Math. 2000;104:45–62.
-
- Akutsu T. Recent advances in RNA secondary structure prediction with pseudoknots. Current Bioinform. 2006;1:115–129.
-
- Andronescu M., et al. Efficient parameter estimation for RNA secondary structure prediction. Bioinformatics. 2007;23:i19–i28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

