Meta-IDBA: a de Novo assembler for metagenomic data
- PMID: 21685107
- PMCID: PMC3117360
- DOI: 10.1093/bioinformatics/btr216
Meta-IDBA: a de Novo assembler for metagenomic data
Abstract
Motivation: Next-generation sequencing techniques allow us to generate reads from a microbial environment in order to analyze the microbial community. However, assembling of a set of mixed reads from different species to form contigs is a bottleneck of metagenomic research. Although there are many assemblers for assembling reads from a single genome, there are no assemblers for assembling reads in metagenomic data without reference genome sequences. Moreover, the performances of these assemblers on metagenomic data are far from satisfactory, because of the existence of common regions in the genomes of subspecies and species, which make the assembly problem much more complicated.
Results: We introduce the Meta-IDBA algorithm for assembling reads in metagenomic data, which contain multiple genomes from different species. There are two core steps in Meta-IDBA. It first tries to partition the de Bruijn graph into isolated components of different species based on an important observation. Then, for each component, it captures the slight variants of the genomes of subspecies from the same species by multiple alignments and represents the genome of one species, using a consensus sequence. Comparison of the performances of Meta-IDBA and existing assemblers, such as Velvet and Abyss for different metagenomic datasets shows that Meta-IDBA can reconstruct longer contigs with similar accuracy.
Availability: Meta-IDBA toolkit is available at our website http://www.cs.hku.hk/~alse/metaidba.
Contact: chin@cs.hku.hk.
Figures


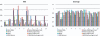
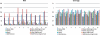

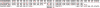
References
-
- Fofanov Y., et al. How independent are the appearances of n-mers in different genomes? Bioinformatics. 2004;20:2421–2428. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

