Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function
- PMID: 21685260
- PMCID: PMC3160020
- DOI: 10.1105/tpc.111.087205
Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function
Abstract
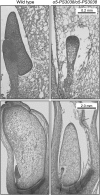
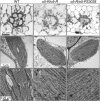
The maize (Zea mays) opaque5 (o5) locus was shown to encode the monogalactosyldiacylglycerol synthase MGD1. Null and point mutations of o5 that affect the vitreous nature of mature endosperm engendered an allelic series of lines with stepwise reductions in gene function. C(18:3)/C(18:2) galactolipid abundance in seedling leaves was reduced proportionally, without significant effects on total galactolipid content. This alteration in polar lipid composition disrupted the organization of thylakoid membranes into granal stacks. Total galactolipid abundance in endosperm was strongly reduced in o5(-) mutants, causing developmental defects and changes in starch production such that the normal simple granules were replaced with compound granules separated by amyloplast membrane. Complete loss of MGD1 function in a null mutant caused kernel lethality owing to failure in both endosperm and embryo development. The data demonstrate that low-abundance galactolipids with five double bonds serve functions in plastid membranes that are not replaced by the predominant species with six double bonds. Furthermore, the data identify a function of amyloplast membranes in the development of starch granules. Finally, the specific changes in lipid composition suggest that MGD1 can distinguish the constituency of acyl groups on its diacylglycerol substrate based upon the degree of desaturation.
Figures








Similar articles
-
The Spatiotemporal Deposition of Lysophosphatidylcholine Within Starch Granules of Maize Endosperm and its Relationships to the Expression of Genes Involved in Endoplasmic Reticulum-Amyloplast Lipid Trafficking and Galactolipid Synthesis.Plant Cell Physiol. 2019 Jan 1;60(1):139-151. doi: 10.1093/pcp/pcy198. Plant Cell Physiol. 2019. PMID: 30295886
-
Inducible knockdown of MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE1 reveals roles of galactolipids in organelle differentiation in Arabidopsis cotyledons.Plant Physiol. 2014 Nov;166(3):1436-49. doi: 10.1104/pp.114.250050. Epub 2014 Sep 24. Plant Physiol. 2014. PMID: 25253888 Free PMC article.
-
A predicted plastid rhomboid protease affects phosphatidic acid metabolism in Arabidopsis thaliana.Plant J. 2019 Sep;99(5):978-987. doi: 10.1111/tpj.14377. Epub 2019 Jun 7. Plant J. 2019. PMID: 31062431 Free PMC article.
-
Green light for galactolipid trafficking.Curr Opin Plant Biol. 2004 Jun;7(3):262-9. doi: 10.1016/j.pbi.2004.03.009. Curr Opin Plant Biol. 2004. PMID: 15134746 Review.
-
Type A and type B monogalactosyldiacylglycerol synthases are spatially and functionally separated in the plastids of higher plants.Plant Physiol Biochem. 2009 Jun;47(6):518-25. doi: 10.1016/j.plaphy.2008.12.012. Epub 2008 Dec 27. Plant Physiol Biochem. 2009. PMID: 19179086 Review.
Cited by
-
Cytosolic malate dehydrogenase 4 modulates cellular energetics and storage reserve accumulation in maize endosperm.Plant Biotechnol J. 2020 Dec;18(12):2420-2435. doi: 10.1111/pbi.13416. Epub 2020 Jun 14. Plant Biotechnol J. 2020. PMID: 32436613 Free PMC article.
-
Maize opaque mutants are no longer so opaque.Plant Reprod. 2018 Sep;31(3):319-326. doi: 10.1007/s00497-018-0344-3. Epub 2018 Jul 5. Plant Reprod. 2018. PMID: 29978299 Free PMC article. Review.
-
Nonredundant function of zeins and their correct stoichiometric ratio drive protein body formation in maize endosperm.Plant Physiol. 2013 Jul;162(3):1359-69. doi: 10.1104/pp.113.218941. Epub 2013 May 15. Plant Physiol. 2013. PMID: 23677936 Free PMC article.
-
Carotenoids modulate kernel texture in maize by influencing amyloplast envelope integrity.Nat Commun. 2020 Oct 22;11(1):5346. doi: 10.1038/s41467-020-19196-9. Nat Commun. 2020. PMID: 33093471 Free PMC article.
-
Deletion mutagenesis identifies a haploinsufficient role for γ-zein in opaque2 endosperm modification.Plant Physiol. 2014 Jan;164(1):119-30. doi: 10.1104/pp.113.230961. Epub 2013 Nov 8. Plant Physiol. 2014. PMID: 24214534 Free PMC article.
References
-
- Andorf C.M., Lawrence C.J., Harper L.C., Schaeffer M.L., Campbell D.A., Sen T.Z. (2010). The Locus Lookup tool at MaizeGDB: Identification of genomic regions in maize by integrating sequence information with physical and genetic maps. Bioinformatics 26: 434–436 - PubMed
-
- Awai K., Maréchal E., Block M.A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2001). Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 10960–10965 - PMC - PubMed
-
- Badenhuizen N.P. (1969). The Biogenesis of Starch Granules in Higher Plants. (New York: Appleton-Century-Crofts; ).
-
- Ball S.G., Morell M.K. (2003). From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54: 207–233 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

